cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma
- PMID: 15380028
- PMCID: PMC520814
- DOI: 10.1186/1471-2164-5-70
cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma
Abstract
Background: Recurrent non-random genomic alterations are the hallmarks of cancer and the characterization of these imbalances is critical to our understanding of tumorigenesis and cancer progression.
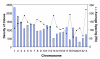
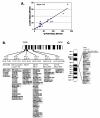
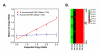
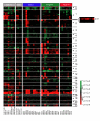
Results: We performed array-comparative genomic hybridization (A-CGH) on cDNA microarrays containing 42,000 elements in neuroblastoma (NB). We found that only two chromosomes (2p and 12q) had gene amplifications and all were in the MYCN amplified samples. There were 6 independent non-contiguous amplicons (10.4-69.4 Mb) on chromosome 2, and the largest contiguous region was 1.7 Mb bounded by NAG and an EST (clone: 757451); the smallest region was 27 Kb including an EST (clone: 241343), NCYM, and MYCN. Using a probabilistic approach to identify single copy number changes, we systemically investigated the genomic alterations occurring in Stage 1 and Stage 4 NBs with and without MYCN amplification (stage 1-, 4-, and 4+). We have not found genomic alterations universally present in all (100%) three subgroups of NBs. However we identified both common and unique patterns of genomic imbalance in NB including gain of 7q32, 17q21, 17q23-24 and loss of 3p21 were common to all three categories. Finally we confirm that the most frequent specific changes in Stage 4+ tumors were the loss of 1p36 with gain of 2p24-25 and they had fewer genomic alterations compared to either stage 1 or 4-, indicating that for this subgroup of poor risk NB requires a smaller number of genomic changes are required to develop the malignant phenotype.
Conclusions: cDNA A-CGH analysis is an efficient method for the detection and characterization of amplicons. Furthermore we were able to detect single copy number changes using our probabilistic approach and identified genomic alterations specific to stage and MYCN amplification.
Figures
References
-
- Brodeur GM, Azar C, Brother M, Hiemstra J, Kaufman B, Marshall H, Moley J, Nakagawara A, Saylors R, Scavarda N, et al. Neuroblastoma. Effect of genetic factors on prognosis and treatment. Cancer. 1992;70:1685–94. - PubMed
-
- Brinkschmidt C, Christiansen H, Terpe HJ, Simon R, Boecker W, Lampert F, Stoerkel S. Comparative genomic hybridization (CGH) analysis of neuroblastomas--an important methodological approach in paediatric tumour pathology. J Pathol. 1997;181:394–400. doi: 10.1002/(SICI)1096-9896(199704)181:4<394::AID-PATH800>3.3.CO;2-T. - DOI - PubMed
-
- Plantaz D, Vandesompele J, Van Roy N, Lastowska M, Bown N, Combaret V, Favrot MC, Delattre O, Michon J, Benard J, et al. Comparative genomichybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer. 2001;91:680–6. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1114>3.0.CO;2-R. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

