A novel strategy for the identification of protein-DNA contacts by photocrosslinking and mass spectrometry
- PMID: 15383647
- PMCID: PMC519130
- DOI: 10.1093/nar/gnh131
A novel strategy for the identification of protein-DNA contacts by photocrosslinking and mass spectrometry
Abstract
Photochemical crosslinking is a method for studying the molecular details of protein-nucleic acid interactions. In this study, we describe a novel strategy to localize crosslinked amino acid residues that combines laser-induced photocrosslinking, proteolytic digestion, Fe3+-IMAC (immobilized metal affinity chromatography) purification of peptide-oligodeoxynucleotide heteroconjugates and hydrolysis of oligodeoxynucleotides by hydrogen fluoride (HF), with efficient matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS). The new method is illustrated by the identification of the DNA-binding site of the restriction endonuclease MboI. Photoactivatable 5-iododeoxyuridine was incorporated into a single site within the DNA recognition sequence (GATC) of MboI. Ultraviolet irradiation of the protein-DNA complex with a helium/cadmium laser at 325 nm resulted in 15% crosslinking yield. Proteolytic digestion with different proteases produced various peptide-oligodeoxynucleotide adducts that were purified together with free oligodeoxynucleotide by Fe3+-IMAC. A combination of MS analysis of the peptide-nucleosides obtained after hydrolysis by HF and their fragmentation by MS/MS revealed that Lys209 of MboI was crosslinked to the MboI recognition site at the position of the adenine, demonstrating that the region around Lys209 is involved in specific binding of MboI to its DNA substrate. This method is suitable for the fast identification of the site of contact between proteins and nucleic acids starting from picomole quantities of crosslinked complexes.
Figures
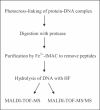

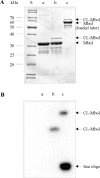
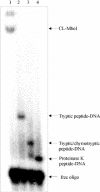
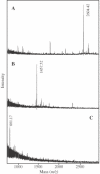

References
-
- Willis M.C., Hicke,B.J., Uhlenbeck,O.C., Cech,T.R. and Koch,T.H. (1993) Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science, 262, 1255–1257. - PubMed
-
- Meisenheimer K.M. and Koch,T.H. (1997) Photocross-linking of nucleic acids to associated proteins. Crit. Rev. Biochem. Mol. Biol., 32, 101–140. - PubMed
-
- Norris C.L., Meisenheimer,P.L. and Koch,T.H. (1996) Mechanistic studies of the 5-iodouracil chromophore relevant to its use in nucleoprotein photocrosslinking. J. Am. Chem. Soc., 118, 5796–5803.
-
- Merrill B.M., Williams,K.R., Chase,J.W. and Konigsberg,W.H. (1984) Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. J. Biol. Chem., 259, 10850–10856. - PubMed
-
- Allen T.D., Wick,K.L. and Matthews,K.S. (1991) Identification of amino acids in lac repressor protein cross-linked to operator DNA specifically substituted with bromodeoxyuridine. J. Biol. Chem., 266, 6113–6119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

