The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes
- PMID: 15383675
- PMCID: PMC1370636
- DOI: 10.1261/rna.7105504
The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes
Abstract
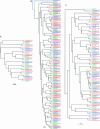
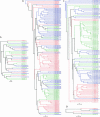
The Drosophila melanogaster Down syndrome cell adhesion molecule (Dscam) gene encodes an axon guidance receptor and can generate 38,016 different isoforms via the alternative splicing of 95 variable exons. Dscam contains 10 immunoglobulin (Ig), six Fibronectin type III, a transmembrane (TM), and cytoplasmic domains. The different Dscam isoforms vary in the amino acid sequence of three of the Ig domains and the TM domain. Here, we have compared the organization of the Dscam gene from three members of the Drosophila subgenus (D. melanogaster, D. pseudoobscura, and D. virilis), the mosquito Anopheles gambiae, and the honeybee Apis mellifera. Each of these organisms contains numerous alternative exons and can potentially synthesize tens of thousands of isoforms. Interestingly, most of the alternative exons in one species are more similar to one another than to the corresponding alternative exons in the other species. These observations provide strong evidence that many of the alternative exons have arisen by reiterative exon duplication and deletion events. In addition, these findings suggest that the expression of a large Dscam repertoire is more important for the development and function of the insect nervous system than the actual sequence of each isoform.
Copyright 2004 RNA Society
Figures


References
-
- Barlow, G.M., Micales, B., Chen, X.N., Lyons, G.E., and Korenberg, J.R. 2002. Mammalian DSCAMs: Roles in the development of the spinal cord, cortex, and cerebellum? Biochem. Biophys. Res. Commun. 293: 881–891. - PubMed
-
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. - PubMed
-
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. - PubMed
-
- Hummel, T., Vasconcelos, M.L., Clemens, J.C., Fishilevich, Y., Vosshall, L.B., and Zipursky, S.L. 2003. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron 37: 221–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
