Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides
- PMID: 15383681
- PMCID: PMC1370650
- DOI: 10.1261/rna.7810204
Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides
Abstract
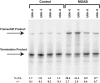
Evidence is presented that morpholino, 2'-O-methyl, phosphorothioate, and RNA antisense oligonucleotides can direct site-specific -1 translational frameshifting when annealed to mRNA downstream from sequences where the P- and A-site tRNAs are both capable of repairing with -1 frame codons. The efficiency of ribosomes shifting into the new frame can be as high as 40%, determined by the sequence of the frameshift site, as well as the location, sequence composition, and modification of the antisense oligonucleotide. These results demonstrate that a perfect duplex formed by complementary oligonucleotides is sufficient to induce high level -1 frameshifting. The implications for the mechanism of action of natural programmed translational frameshift stimulators are discussed.
Copyright 2004 RNA Society
Figures






References
-
- Agrawal, R.K., Heagle, A.B., Penczek, P., Grassucci, R.A., and Frank, J. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6: 643–647. - PubMed
-
- Baranov, P.V., Gesteland, R.F., and Atkins, J.F. 2002. Recoding: Translational bifurcations in gene expression. Gene 286: 187–201. - PubMed
-
- Brierley, I. and Pennell, S. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic -1 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 66: 233–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
