Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function
- PMID: 15385536
- PMCID: PMC1201109
- DOI: 10.1074/jbc.M409421200
Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function
Abstract
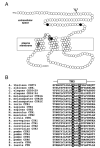
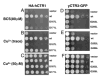
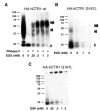
Members of the copper uptake transporter (CTR) family from yeast, plants, and mammals including human are required for cellular uptake of the essential metal copper. Based on biochemical data, CTRs have three transmembrane domains and have been shown to oligomerize in the membrane. Among individual members of the family, there is little amino acid sequence identity, raising questions as to how these proteins adopt a common fold, oligomerize, and participate in copper transport. Using site-directed mutagenesis, tryptophan scanning, genetic complementation, subcellular localization, chemical cross-linking, and the yeast unfolded protein response, we demonstrated that at least half of the third transmembrane domain (TM3) plays a vital role in CTR structure and function. The results of our analysis showed that TM3 contains two functionally distinct faces. One face bears a highly conserved Gly-X-X-X-Gly (GG4) motif, which we showed to be essential for CTR oligomerization. Moreover, we showed that steric constraints reach past the GG4-motif itself including amino acid residues that are not conserved throughout the CTR family. A second face of TM3 contains three amino acid positions that, when mutated to tryptophan, cause predominantly abnormal localization but are still partially functional in growth complementation experiments. These mutations cluster on the face opposite to the GG4-bearing face of TM3 where they may mediate interactions with the remaining two transmembrane domains. Taken together, our data support TM3 as being buried within trimeric CTR where it plays an essential role in CTR assembly.
Figures




References
-
- Linder, M. C. (1991) Biochemistry of Copper, Plenum Press, New York
-
- Pena MM, Lee J, Thiele DJ. J Nutr. 1999;129:1251–1260. - PubMed
-
- DiDonato M, Sarkar B. Biochim Biophys Acta. 1997;1360:3–16. - PubMed
-
- Schaefer M, Gitlin JD. Am J Physiol. 1999;276:G311–G314. - PubMed
-
- Strausak D, Mercer JF, Dieter HH, Stremmel W, Multhaup G. Brain Res Bull. 2001;55:175–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

