Coordinating assembly and export of complex bacterial proteins
- PMID: 15385959
- PMCID: PMC524343
- DOI: 10.1038/sj.emboj.7600409
Coordinating assembly and export of complex bacterial proteins
Abstract
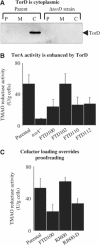
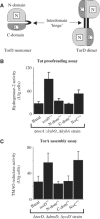
The Escherichia coli twin-arginine protein transport (Tat) system is a molecular machine dedicated to the translocation of fully folded substrate proteins across the energy-transducing inner membrane. Complex cofactor-containing Tat substrates, such as the model (NiFe) hydrogenase-2 and trimethylamine N-oxide reductase (TorA) systems, acquire their redox cofactors prior to export from the cell and require to be correctly assembled before transport can proceed. It is likely, therefore, that cellular mechanisms exist to prevent premature export of immature substrates. Using a combination of genetic and biochemical approaches including gene knockouts, signal peptide swapping, complementation, and site-directed mutagenesis, we highlight here this crucial 'proofreading' or 'quality control' activity in operation during assembly of complex endogenous Tat substrates. Our experiments successfully uncouple the Tat transport and cofactor-insertion activities of the TorA-specific chaperone TorD and demonstrate unequivocally that TorD recognises the TorA twin-arginine signal peptide. It is proposed that some Tat signal peptides operate in tandem with cognate binding chaperones to orchestrate the assembly and transport of complex enzymes.
Figures


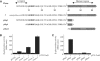


Similar articles
-
Intrinsic GTPase activity of a bacterial twin-arginine translocation proofreading chaperone induced by domain swapping.FEBS J. 2010 Jan;277(2):511-25. doi: 10.1111/j.1742-4658.2009.07507.x. FEBS J. 2010. PMID: 20064164
-
Sequence analysis of bacterial redox enzyme maturation proteins (REMPs).Can J Microbiol. 2004 Apr;50(4):225-38. doi: 10.1139/w03-117. Can J Microbiol. 2004. PMID: 15213747 Review.
-
Chaperones involved in assembly and export of N-oxide reductases.Biochem Soc Trans. 2005 Feb;33(Pt 1):124-6. doi: 10.1042/BST0330124. Biochem Soc Trans. 2005. PMID: 15667282
-
Characterization of a pre-export enzyme-chaperone complex on the twin-arginine transport pathway.Biochem J. 2013 May 15;452(1):57-66. doi: 10.1042/BJ20121832. Biochem J. 2013. PMID: 23452237 Free PMC article.
-
Assembly of membrane-bound respiratory complexes by the Tat protein-transport system.Arch Microbiol. 2002 Aug;178(2):77-84. doi: 10.1007/s00203-002-0434-2. Epub 2002 May 22. Arch Microbiol. 2002. PMID: 12115052 Review.
Cited by
-
A synthetic system for expression of components of a bacterial microcompartment.Microbiology (Reading). 2013 Nov;159(Pt 11):2427-2436. doi: 10.1099/mic.0.069922-0. Epub 2013 Sep 6. Microbiology (Reading). 2013. PMID: 24014666 Free PMC article.
-
Exploring the active site of the tungsten, iron-sulfur enzyme acetylene hydratase.J Bacteriol. 2011 Mar;193(5):1229-36. doi: 10.1128/JB.01057-10. Epub 2010 Dec 30. J Bacteriol. 2011. PMID: 21193613 Free PMC article.
-
The twin-arginine translocation (Tat) protein export pathway.Nat Rev Microbiol. 2012 Jun 11;10(7):483-96. doi: 10.1038/nrmicro2814. Nat Rev Microbiol. 2012. PMID: 22683878 Review.
-
A signal sequence suppressor mutant that stabilizes an assembled state of the twin arginine translocase.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):E1958-E1967. doi: 10.1073/pnas.1615056114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223511 Free PMC article.
-
Biosynthesis of selenate reductase in Salmonella enterica: critical roles for the signal peptide and DmsD.Microbiology (Reading). 2016 Dec;162(12):2136-2146. doi: 10.1099/mic.0.000381. Epub 2016 Oct 20. Microbiology (Reading). 2016. PMID: 27902441 Free PMC article.
References
-
- Alami M, Luke I, Deitermann S, Eisner G, Koch HG, Brunner J, Müller M (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 12: 937–946 - PubMed
-
- Berks BC, Palmer T, Sargent F (2003) The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol 47: 187–254 - PubMed
-
- Blasco F, Dos Santos JP, Magalon A, Frixon C, Guigliarelli B, Santini CL, Giordano G (1998) NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol Microbiol 28: 435–447 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

