Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity
- PMID: 15385962
- PMCID: PMC524339
- DOI: 10.1038/sj.emboj.7600402
Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity
Abstract
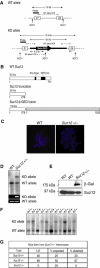
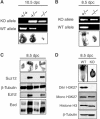
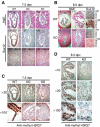
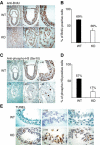
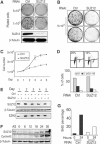
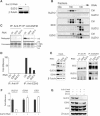
SUZ12 is a recently identified Polycomb group (PcG) protein, which together with EZH2 and EED forms different Polycomb repressive complexes (PRC2/3). These complexes contain histone H3 lysine (K) 27/9 and histone H1 K26 methyltransferase activity specified by the EZH2 SET domain. Here we show that mice lacking Suz12, like Ezh2 and Eed mutant mice, are not viable and die during early postimplantation stages displaying severe developmental and proliferative defects. Consistent with this, we demonstrate that SUZ12 is required for proliferation of cells in tissue culture. Furthermore, we demonstrate that SUZ12 is essential for the activity and stability of the PRC2/3 complexes in mouse embryos, in tissue culture cells and in vitro. Strikingly, Suz12-deficient embryos show a specific loss of di- and trimethylated H3K27, demonstrating that Suz12 is indeed essential for EZH2 activity in vivo. In conclusion, our data demonstrate an essential role of SUZ12 in regulating the activity of the PRC2/3 complexes, which are required for regulating proliferation and embryogenesis.
Figures

References
-
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Muller J (2001) Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128: 3371–3379 - PubMed
-
- Brock HW, van Lohuizen M (2001) The Polycomb group—no longer an exclusive club? Curr Opin Genet Dev 11: 175–181 - PubMed
-
- Brummelkamp TR, Bernards R, Agami R (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247 - PubMed
-
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

