VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis
- PMID: 15388450
- PMCID: PMC521886
- DOI: 10.1128/AAC.48.10.3892-3904.2004
VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis
Abstract
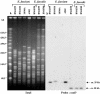
Enterococcus faecium clinical isolates A902 and BM4538, which were resistant to relatively high levels of vancomycin (128 and 64 microg/ml, respectively) and to low levels of teicoplanin (4 microg/ml), and Enterococcus faecalis clinical isolates BM4539 and BM4540, which were resistant to moderate levels of vancomycin (16 microg/ml) and susceptible to teicoplanin (0.25 microg/ml), were studied. They were constitutively resistant by synthesis of peptidoglycan precursors ending with d-alanyl-d-lactate and harbored a chromosomal vanD gene cluster which was not transferable by conjugation to other enterococci. VanX(D) activity, which is not required in the absence of d-Ala-d-Ala, was low in the four strains, although none of the conserved residues was mutated; and the constitutive VanY(D) activity in the membrane fractions was inhibited by penicillin G. The mutations E(13)G in the region of d-alanine:d-alanine ligase (which is implicated in d-Ala1 binding in A902) and S(319)N of the serine involved in ATP binding in BM4538 and a 7-bp insertion at different locations in BM4539 and BM4540 (which led to putative truncated proteins) led to the production of an impaired enzyme and accounted for the lack of d-Ala-d-Ala-containing peptidoglycan precursors. The same 7-bp insertion in vanS(D) of BM4539 and BM4540 and a 1-bp deletion in vanS(D) of A902, which in each case led to a putative truncated and presumably nonfunctional protein, could account for the constitutive resistance. Strain BM4538, with a functional VanS(D), had a G(140)E mutation in VanR(D) that could be responsible for constitutive glycopeptide resistance. This would represent the first example of constitutive van gene expression due to a mutation in the structural gene for a VanR transcriptional activator. Study of these four additional strains that could be distinguished on the basis of their various assortments of mutations confirmed that all VanD-type strains isolated so far have mutations in the ddl housekeeping gene and in the acquired vanS(D) or vanR(D) gene that lead to constitutive resistance to vancomycin.
Figures




References
-
- Aiba, H., F. Nakasai, S. Mizushima, and T. Mizuno. 1989. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J. Biol. Chem. 264:14090-14094. - PubMed
-
- Arthur, M., F. Depardieu, L. Cabanié, P. Reynolds, and P. Courvalin. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819-830. - PubMed
-
- Arthur, M., F. Depardieu, and P. Courvalin. 1999. Regulated interactions between partner and non partner sensors and responses regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145:1849-1858. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

