Sequence composition and genome organization of maize
- PMID: 15388850
- PMCID: PMC521949
- DOI: 10.1073/pnas.0406163101
Sequence composition and genome organization of maize
Abstract
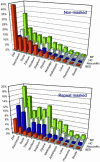
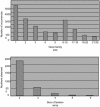
Zea mays L. ssp. mays, or corn, one of the most important crops and a model for plant genetics, has a genome approximately 80% the size of the human genome. To gain global insight into the organization of its genome, we have sequenced the ends of large insert clones, yielding a cumulative length of one-eighth of the genome with a DNA sequence read every 6.2 kb, thereby describing a large percentage of the genes and transposable elements of maize in an unbiased approach. Based on the accumulative 307 Mb of sequence, repeat sequences occupy 58% and genic regions occupy 7.5%. A conservative estimate predicts approximately 59,000 genes, which is higher than in any other organism sequenced so far. Because the sequences are derived from bacterial artificial chromosome clones, which are ordered in overlapping bins, tagged genes are also ordered along continuous chromosomal segments. Based on this positional information, roughly one-third of the genes appear to consist of tandemly arrayed gene families. Although the ancestor of maize arose by tetraploidization, fewer than half of the genes appear to be present in two orthologous copies, indicating that the maize genome has undergone significant gene loss since the duplication event.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

