Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan
- PMID: 15447792
- PMCID: PMC523852
- DOI: 10.1186/1471-2350-5-24
Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan
Abstract
Background: Mutant alleles of TMPRSS3 are associated with nonsyndromic recessive deafness (DFNB8/B10). TMPRSS3 encodes a predicted secreted serine protease, although the deduced amino acid sequence has no signal peptide. In this study, we searched for mutant alleles of TMPRSS3 in families from Pakistan and Newfoundland with recessive deafness co-segregating with DFNB8/B10 linked haplotypes and also more thoroughly characterized the genomic structure of TMPRSS3.
Methods: We enrolled families segregating recessive hearing loss from Pakistan and Newfoundland. Microsatellite markers flanking the TMPRSS3 locus were used for linkage analysis. DNA samples from participating individuals were sequenced for TMPRSS3. The structure of TMPRSS3 was characterized bioinformatically and experimentally by sequencing novel cDNA clones of TMPRSS3.
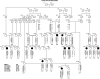
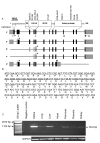
Results: We identified mutations in TMPRSS3 in four Pakistani families with recessive, nonsyndromic congenital deafness. We also identified two recessive mutations, one of which is novel, of TMPRSS3 segregating in a six-generation extended family from Newfoundland. The spectrum of TMPRSS3 mutations is reviewed in the context of a genotype-phenotype correlation. Our study also revealed a longer isoform of TMPRSS3 with a hitherto unidentified exon encoding a signal peptide, which is expressed in several tissues.
Conclusion: Mutations of TMPRSS3 contribute to hearing loss in many communities worldwide and account for 1.8% (8 of 449) of Pakistani families segregating congenital deafness as an autosomal recessive trait. The newly identified TMPRSS3 isoform e will be helpful in the functional characterization of the full length protein.
Figures

References
-
- Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. - PubMed
-
- Ben-Yosef T, Wattenhofer M, Riazuddin S, Ahmed ZM, Scott HS, Kudoh J, Shibuya K, Antonarakis SE, Bonne-Tamir B, Radhakrishna U, Naz S, Ahmed Z, Pandya A, Nance WE, Wilcox ER, Friedman TB, Morell RJ. Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J Med Genet. 2001;38:396–400. doi: 10.1136/jmg.38.6.396. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

