Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis
- PMID: 15448182
- PMCID: PMC521639
- DOI: 10.1093/nar/gkh793
Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis
Abstract
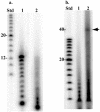
A basic problem in gene synthesis is the acquisition of many short oligonucleotide sequences needed for the assembly of genes. Photolithographic methods for the massively parallel synthesis of high-density oligonucleotide arrays provides a potential source, once appropriate methods have been devised for their elution in forms suitable for enzyme-catalyzed assembly. Here, we describe a method based on the photolithographic synthesis of long (>60mers) single-stranded oligonucleotides, using a modified maskless array synthesizer. Once the covalent bond between the DNA and the glass surface is cleaved, the full-length oligonucleotides are selected and amplified using PCR. After cleavage of flanking primer sites, a population of unique, internal 40mer dsDNA sequences are released and are ready for use in biological applications. Subsequent gene assembly experiments using this DNA pool were performed and were successful in creating longer DNA fragments. This is the first report demonstrating the use of eluted chip oligonucleotides in biological applications such as PCR and assembly PCR.
Figures




References
-
- Cello J., Paul,A.V. and Wimmer,E. (2002) Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science, 297, 1016–1018. - PubMed
-
- Pirrung M.C., Wang,L. and Montague-Smith,M.P. (2001) 3′-nitrophenylpropyloxycarbonyl (NPPOC) protecting groups for high-fidelity automated 5′ → 3′ photochemical DNA synthesis. Org. Lett., 3, 1105–1108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

