A comparative method for finding and folding RNA secondary structures within protein-coding regions
- PMID: 15448187
- PMCID: PMC519121
- DOI: 10.1093/nar/gkh839
A comparative method for finding and folding RNA secondary structures within protein-coding regions
Abstract
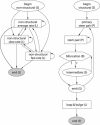
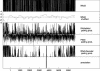
Existing computational methods for RNA secondary-structure prediction tacitly assume RNA to only encode functional RNA structures. However, experimental studies have revealed that some RNA sequences, e.g. compact viral genomes, can simultaneously encode functional RNA structures as well as proteins, and evidence is accumulating that this phenomenon may also be found in Eukaryotes. We here present the first comparative method, called RNA-DECODER, which explicitly takes the known protein-coding context of an RNA-sequence alignment into account in order to predict evolutionarily conserved secondary-structure elements, which may span both coding and non-coding regions. RNA-DECODER employs a stochastic context-free grammar together with a set of carefully devised phylogenetic substitution-models, which can disentangle and evaluate the different kinds of overlapping evolutionary constraints which arise. We show that RNA-DECODER's parameters can be automatically trained to successfully fold known secondary structures within the HCV genome. We scan the genomes of HCV and polio virus for conserved secondary-structure elements, and analyze performance as a function of available evolutionary information. On known secondary structures, RNA-DECODER shows a sensitivity similar to the programs MFOLD, PFOLD and RNAALIFOLD. When scanning the entire genomes of HCV and polio virus for structure elements, RNA-DECODER's results indicate a markedly higher specificity than MFOLD, PFOLD and RNAALIFOLD.
Figures



Similar articles
-
An evolutionary model for protein-coding regions with conserved RNA structure.Mol Biol Evol. 2004 Oct;21(10):1913-22. doi: 10.1093/molbev/msh199. Epub 2004 Jun 30. Mol Biol Evol. 2004. PMID: 15229291
-
Pfold: RNA secondary structure prediction using stochastic context-free grammars.Nucleic Acids Res. 2003 Jul 1;31(13):3423-8. doi: 10.1093/nar/gkg614. Nucleic Acids Res. 2003. PMID: 12824339 Free PMC article.
-
Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs.Nucleic Acids Res. 2005 Nov 7;33(19):6338-48. doi: 10.1093/nar/gki923. Print 2005. Nucleic Acids Res. 2005. PMID: 16275783 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
In silico methods for co-transcriptional RNA secondary structure prediction and for investigating alternative RNA structure expression.Methods. 2017 May 1;120:3-16. doi: 10.1016/j.ymeth.2017.04.009. Epub 2017 Apr 20. Methods. 2017. PMID: 28433606 Review.
Cited by
-
Random generation of RNA secondary structures according to native distributions.Algorithms Mol Biol. 2011 Oct 12;6:24. doi: 10.1186/1748-7188-6-24. Algorithms Mol Biol. 2011. PMID: 21992500 Free PMC article.
-
Towards Long-Range RNA Structure Prediction in Eukaryotic Genes.Genes (Basel). 2018 Jun 15;9(6):302. doi: 10.3390/genes9060302. Genes (Basel). 2018. PMID: 29914113 Free PMC article. Review.
-
Evidence of a novel RNA secondary structure in the coding region of HIV-1 pol gene.RNA. 2008 Dec;14(12):2478-88. doi: 10.1261/rna.1252608. Epub 2008 Oct 30. RNA. 2008. PMID: 18974280 Free PMC article.
-
Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses.Nucleic Acids Res. 2008 May;36(8):2530-46. doi: 10.1093/nar/gkn096. Epub 2008 Mar 4. Nucleic Acids Res. 2008. PMID: 18319285 Free PMC article.
-
Knotify_V2.0: Deciphering RNA Secondary Structures with H-Type Pseudoknots and Hairpin Loops.Genes (Basel). 2024 May 23;15(6):670. doi: 10.3390/genes15060670. Genes (Basel). 2024. PMID: 38927606 Free PMC article.
References
-
- Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. - PubMed
-
- Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. - PubMed
-
- Xiang W., Paul,A.V. and Wimmer,E. (1997) RNA signals in entero- and rhinovirus genome replication. Semin. Virol., 8, 256–273.
-
- Huthoff H. and Berkhout,B. (2002) Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry, 41, 10439–10445. - PubMed

