Local graph alignment and motif search in biological networks
- PMID: 15448202
- PMCID: PMC522014
- DOI: 10.1073/pnas.0305199101
Local graph alignment and motif search in biological networks
Abstract
Interaction networks are of central importance in postgenomic molecular biology, with increasing amounts of data becoming available by high-throughput methods. Examples are gene regulatory networks or protein interaction maps. The main challenge in the analysis of these data is to read off biological functions from the topology of the network. Topological motifs, i.e., patterns occurring repeatedly at different positions in the network, have recently been identified as basic modules of molecular information processing. In this article, we discuss motifs derived from families of mutually similar but not necessarily identical patterns. We establish a statistical model for the occurrence of such motifs, from which we derive a scoring function for their statistical significance. Based on this scoring function, we develop a search algorithm for topological motifs called graph alignment, a procedure with some analogies to sequence alignment. The algorithm is applied to the gene regulation network of Escherichia coli.
Figures

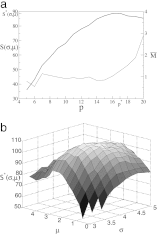
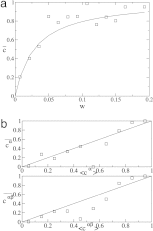

References
-
- Durbin, R., Eddy, S. R., Krogh, A. & Mitchison, G. (1998) Biological Sequence Analysis (Cambridge Univ. Press, Cambridge, U.K.).
-
- Davidson, E. H. (2001) Genomic Regulatory Systems: Development and Evolution (Academic, San Diego).
-
- Tautz, D. (2000) Curr. Opin. Genet. Dev. 1 575–579. - PubMed
-
- Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403 623–627. - PubMed
-
- Lockhart, D. J. & Winzeler, E. A. (2000) Nature 405 827–836. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

