Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker
- PMID: 15448206
- PMCID: PMC521937
- DOI: 10.1073/pnas.0405154101
Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker
Abstract
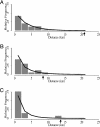
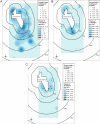
Sampling methods and results of a gene flow study are described that will be of interest to plant scientists, evolutionary biologists, ecologists, and stakeholders assessing the environmental safety of transgenic crops. This study documents gene flow on a landscape level from creeping bentgrass (Agrostis stolonifera L.), one of the first wind-pollinated, perennial, and highly outcrossing transgenic crops being developed for commercial use. Most of the gene flow occurred within 2 km in the direction of prevailing winds. The maximal gene flow distances observed were 21 km and 14 km in sentinel and resident plants, respectively, that were located in primarily nonagronomic habitats. The selectable marker used in these studies was the CP4 EPSPS gene derived from Agrobacterium spp. strain CP4 that encodes 5-enol-pyruvylshikimate-3-phosphate synthase and confers resistance to glyphosate herbicide. Evidence for gene flow to 75 of 138 sentinel plants of A. stolonifera and to 29 of 69 resident Agrostis plants was based on seedling progeny survival after spraying with glyphosate in greenhouse assays and positive TraitChek, PCR, and sequencing results. Additional studies are needed to determine whether introgression will occur and whether it will affect the ecological fitness of progeny or the structure of plant communities in which transgenic progeny may become established.
Figures


References
-
- Ellstrand, N. C. (2003) Dangerous Liasions? When Cultivated Plants Mate With Wild Relatives (John Hopkins Univ. Press, Baltimore).
-
- Klinger, T., Elam, D. R. & Ellstrand, N. C. (1991) Conserv. Biol. 5, 531-535.
-
- Arias, D. M. & Rieseberg, L. H. (1994) Theor. Appl. Genet. 89, 665-660. - PubMed
-
- Kirkpatrick, K. J. & Wilson, H. D. (1988) Am. J. Botany 75, 519-527.
-
- Reiger, M. A., Lamond, M., Preston, C., Powles, S. B. & Roush, R. T. (2002) Science 296, 2386-2388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

