3D image segmentation of deformable objects with joint shape-intensity prior models using level sets
- PMID: 15450223
- PMCID: PMC2832842
- DOI: 10.1016/j.media.2004.06.008
3D image segmentation of deformable objects with joint shape-intensity prior models using level sets
Abstract
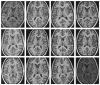
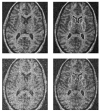
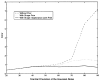
We propose a novel method for 3D image segmentation, where a Bayesian formulation, based on joint prior knowledge of the object shape and the image gray levels, along with information derived from the input image, is employed. Our method is motivated by the observation that the shape of an object and the gray level variation in an image have consistent relations that provide configurations and context that aid in segmentation. We define a maximum a posteriori (MAP) estimation model using the joint prior information of the object shape and the image gray levels to realize image segmentation. We introduce a representation for the joint density function of the object and the image gray level values, and define a joint probability distribution over the variations of the object shape and the gray levels contained in a set of training images. By estimating the MAP shape of the object, we formulate the shape-intensity model in terms of level set functions as opposed to landmark points of the object shape. In addition, we evaluate the performance of the level set representation of the object shape by comparing it with the point distribution model (PDM). We found the algorithm to be robust to noise and able to handle multidimensional data, while able to avoid the need for explicit point correspondences during the training phase. Results and validation from various experiments on 2D and 3D medical images are shown.
Figures






References
-
- Caselles V, Catte F, Coll T, Dibos F. A geometric model for active contours in image processing. Numer. Math. 1993;66:1–31.
-
- Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int. J. Comput. Vis. 1997;22(1):61–79.
-
- Chan T, Vese L. Active contours without edges. IEEE Trans. Image Process. 2001;10(2):266–277. - PubMed
-
- Chen Y, Tagare H, Thiruvenkadam S, Huang F, Wilson D, Gopinath KS, Briggs RW, Geiser EA. Using prior shapes in geometric active contours in a variational framework. Int. J. Comput. Vis. 2002;50(3):315–328.
-
- Cootes T, Beeston C, Edwards G, Taylor C. A unified framework for atlas matching using active appearance models. Inf. Process. Med. Imaging (IPMI) 1999
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

