Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase
- PMID: 15452123
- PMCID: PMC1351161
- DOI: 10.1074/jbc.M408573200
Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase
Abstract
Retroviruses utilize cellular dNTPs to perform proviral DNA synthesis in infected host cells. Unlike oncoretroviruses, which replicate in dividing cells, lentiviruses, such as human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus, are capable of efficiently replicating in non-dividing cells (terminally differentiated macrophages) as well as dividing cells (i.e. activated CD4+ T cells). In general, non-dividing cells are likely to have low cellular dNTP content compared with dividing cells. Here, by employing a novel assay for cellular dNTP content, we determined the dNTP concentrations in two HIV-1 target cells, macrophages and activated CD4+ T cells. We found that human macrophages contained 130-250-fold lower dNTP concentrations than activated human CD4+ T cells. Biochemical analysis revealed that, unlike oncoretroviral reverse transcriptases (RTs), lentiviral RTs efficiently synthesize DNA even in the presence of the low dNTP concentrations equivalent to those found in macrophages. In keeping with this observation, HIV-1 vectors containing mutant HIV-1 RTs, which kinetically mimic oncoretroviral RTs, failed to transduce human macrophages despite retaining normal infectivity for activated CD4+ T cells and other dividing cells. These results suggest that the ability of HIV-1 to infect macrophages, which is essential to establishing the early pathogenesis of HIV-1 infection, depends, at least in part, on enzymatic adaptation of HIV-1 RT to efficiently catalyze DNA synthesis in limited cellular dNTP substrate environments.
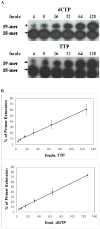
Figures






Similar articles
-
Leishmania induces survival, proliferation and elevated cellular dNTP levels in human monocytes promoting acceleration of HIV co-infection.PLoS Pathog. 2012;8(4):e1002635. doi: 10.1371/journal.ppat.1002635. Epub 2012 Apr 5. PLoS Pathog. 2012. PMID: 22496656 Free PMC article.
-
Mechanistic and Kinetic Differences between Reverse Transcriptases of Vpx Coding and Non-coding Lentiviruses.J Biol Chem. 2015 Dec 11;290(50):30078-86. doi: 10.1074/jbc.M115.691576. Epub 2015 Oct 19. J Biol Chem. 2015. PMID: 26483545 Free PMC article.
-
Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations.J Biol Chem. 2006 May 12;281(19):13388-13395. doi: 10.1074/jbc.M600291200. Epub 2006 Feb 22. J Biol Chem. 2006. PMID: 16497663
-
Intracellular nucleotide levels and the control of retroviral infections.Virology. 2013 Feb 20;436(2):247-54. doi: 10.1016/j.virol.2012.11.010. Epub 2012 Dec 20. Virology. 2013. PMID: 23260109 Free PMC article. Review.
-
Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases.Prog Nucleic Acid Res Mol Biol. 2002;71:91-147. doi: 10.1016/s0079-6603(02)71042-8. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102562 Review.
Cited by
-
Pre-steady state kinetic analysis of HIV-1 reverse transcriptase for non-canonical ribonucleoside triphosphate incorporation and DNA synthesis from ribonucleoside-containing DNA template.Antiviral Res. 2015 Mar;115:75-82. doi: 10.1016/j.antiviral.2014.12.016. Epub 2014 Dec 31. Antiviral Res. 2015. PMID: 25557601 Free PMC article.
-
The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus.J Virol. 2012 Dec;86(23):12552-60. doi: 10.1128/JVI.01657-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973040 Free PMC article.
-
HIV-1 inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3.PLoS One. 2010 Jul 22;5(7):e11733. doi: 10.1371/journal.pone.0011733. PLoS One. 2010. PMID: 20661303 Free PMC article.
-
Mechanism of allosteric activation of SAMHD1 by dGTP.Nat Struct Mol Biol. 2013 Nov;20(11):1304-9. doi: 10.1038/nsmb.2692. Epub 2013 Oct 20. Nat Struct Mol Biol. 2013. PMID: 24141705 Free PMC article.
-
Comparative analysis of the capacity of elite suppressor CD4+ and CD8+ T cells to inhibit HIV-1 replication in monocyte-derived macrophages.J Virol. 2014 Sep 1;88(17):9789-98. doi: 10.1128/JVI.00860-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942573 Free PMC article.
References
-
- Traut TW. Mol Cell Biochem. 1994;140:1–22. - PubMed
-
- Angus SP, Wheeler LJ, Ranmal SA, Zhang X, Markey MP, Mathews CK, Knudsen ES. J Biol Chem. 2002;277:44376–44384. - PubMed
-
- Jackson RC, Lui MS, Boritzki TJ, Morris HP, Weber G. Cancer Res. 1980;40:1286–291. - PubMed
-
- Hauschka PV. Methods Cell Biol. 1973;7:361–462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI49815/AI/NIAID NIH HHS/United States
- S11 NS43499/NS/NINDS NIH HHS/United States
- AI49781/AI/NIAID NIH HHS/United States
- T32 AI049815/AI/NIAID NIH HHS/United States
- R01 AI049781/AI/NIAID NIH HHS/United States
- AI49057/AI/NIAID NIH HHS/United States
- R01 GM049573/GM/NIGMS NIH HHS/United States
- GM49573/GM/NIGMS NIH HHS/United States
- R01 AI087508/AI/NIAID NIH HHS/United States
- R56 AI049057/AI/NIAID NIH HHS/United States
- P01 MH64570/MH/NIMH NIH HHS/United States
- R01 AI049057/AI/NIAID NIH HHS/United States
- P01 MH064570/MH/NIMH NIH HHS/United States
- R56 AI049781/AI/NIAID NIH HHS/United States
- S11 NS043499/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

