VP7 mediates the interaction of rotaviruses with integrin alphavbeta3 through a novel integrin-binding site
- PMID: 15452204
- PMCID: PMC521812
- DOI: 10.1128/JVI.78.20.10839-10847.2004
VP7 mediates the interaction of rotaviruses with integrin alphavbeta3 through a novel integrin-binding site
Abstract
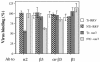
Rotavirus entry is a complex multistep process that depends on the trypsin cleavage of the virus spike protein VP4 into polypeptides VP5 and VP8 and on the interaction of these polypeptides and of VP7, the second viral surface protein, with several cell surface molecules, including integrin alphavbeta3. We characterized the effect of the trypsin cleavage of VP4 on the binding to MA104 cells of the sialic acid-dependent virus strain RRV and its sialic acid-independent variant, nar3. We found that, although the trypsin treatment did not affect the attachment of these viruses to the cell surface, their binding was qualitatively different. In contrast to the trypsin-treated viruses, which initially bound to the cell surface through VP4, the non-trypsin-treated variant nar3 bound to the cell through VP7. Amino acid sequence comparison of the surface proteins of rotavirus and hantavirus, both of which interact with integrin alphavbeta3 in an RGD-independent manner, identified a region shared by rotavirus VP7 and hantavirus G1G2 protein in which six of nine amino acids are identical. This region, which is highly conserved among the VP7 proteins of different rotavirus strains, mediates the binding of rotaviruses to integrin alphavbeta3 and probably represents a novel binding motif for this integrin.
Figures

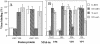

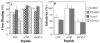



Similar articles
-
Integrin-using rotaviruses bind alpha2beta1 integrin alpha2 I domain via VP4 DGE sequence and recognize alphaXbeta2 and alphaVbeta3 by using VP7 during cell entry.J Virol. 2003 Sep;77(18):9969-78. doi: 10.1128/jvi.77.18.9969-9978.2003. J Virol. 2003. PMID: 12941907 Free PMC article.
-
Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5.J Virol. 2003 Jul;77(13):7254-60. doi: 10.1128/jvi.77.13.7254-7260.2003. J Virol. 2003. PMID: 12805424 Free PMC article.
-
The VP5 domain of VP4 can mediate attachment of rotaviruses to cells.J Virol. 2000 Jan;74(2):593-9. doi: 10.1128/jvi.74.2.593-599.2000. J Virol. 2000. PMID: 10623720 Free PMC article.
-
Molecular biology of rotavirus cell entry.Arch Med Res. 2002 Jul-Aug;33(4):356-61. doi: 10.1016/s0188-4409(02)00374-0. Arch Med Res. 2002. PMID: 12234525 Review.
-
Early events of rotavirus infection: the search for the receptor(s).Novartis Found Symp. 2001;238:47-60; discussion 60-3. doi: 10.1002/0470846534.ch4. Novartis Found Symp. 2001. PMID: 11444034 Review.
Cited by
-
Why cellular stress suppresses adipogenesis in skeletal tissue, but is ineffective in adipose tissue: control of mesenchymal cell differentiation via integrin binding sites in extracellular matrices.Matrix Biol. 2013 Oct-Nov;32(7-8):365-71. doi: 10.1016/j.matbio.2013.06.001. Epub 2013 Jun 18. Matrix Biol. 2013. PMID: 23792045 Free PMC article.
-
The spike protein VP4 defines the endocytic pathway used by rotavirus to enter MA104 cells.J Virol. 2013 Feb;87(3):1658-63. doi: 10.1128/JVI.02086-12. Epub 2012 Nov 21. J Virol. 2013. PMID: 23175367 Free PMC article.
-
Cell surface heat shock protein-mediated entry of tumor cell-adapted rotavirus into U-937 cells.Folia Microbiol (Praha). 2021 Aug;66(4):623-638. doi: 10.1007/s12223-020-00845-x. Epub 2021 May 5. Folia Microbiol (Praha). 2021. PMID: 33950511
-
Assessing the oncolytic potential of rotavirus on mouse myeloma cell line Sp2/0-Ag14.Biomedica. 2020 Jun 15;40(2):362-381. doi: 10.7705/biomedica.4916. Biomedica. 2020. PMID: 32673463 Free PMC article.
-
Ins and Outs of Reovirus: Vesicular Trafficking in Viral Entry and Egress.Trends Microbiol. 2021 Apr;29(4):363-375. doi: 10.1016/j.tim.2020.09.004. Epub 2020 Sep 29. Trends Microbiol. 2021. PMID: 33008713 Free PMC article. Review.
References
-
- Arias, C. F., C. A. Guerrero, E. Mendez, S. Zárate, P. Isa, R. Espinosa, P. Romero, and S. Lopez. 2001. Early events of rotavirus infection: the search for the receptor(s). John Wiley & Sons, Inc., New York, N.Y. - PubMed
-
- Arias, C. F., M. Lizano, and S. López. 1987. Synthesis in Escherichia coli and immunological characterization of a polypeptide containing the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. J. Gen. Virol. 68:633-642. - PubMed
-
- Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

