Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene
- PMID: 15452209
- PMCID: PMC521837
- DOI: 10.1128/JVI.78.20.10888-10905.2004
Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene
Abstract
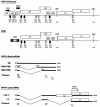
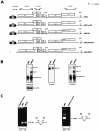
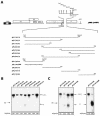
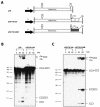
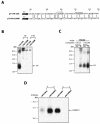
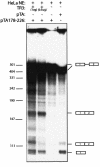
We have previously identified cis-acting RNA sequences in the human papillomavirus type 16 (HPV-16) L1 coding region which inhibit expression of L1 from eukaryotic expression plasmids. Here we have determined the function of one of these RNA elements, and we provide evidence that this RNA element is a splicing silencer which suppresses the use of the 3' splice site located immediately upstream of the L1 AUG. We also show that this splice site is inefficiently utilized as a result of a suboptimal polypyrimidine tract. Introduction of point mutations in the L1 coding region that altered the RNA sequence without affecting the L1 protein sequence resulted in the inactivation of the splicing silencer and induced splicing to the L1 3' splice site. These mutations also prevented the interaction of the RNA silencer with a 35-kDa cellular protein identified here as hnRNP A1. The splicing silencer in L1 inhibits splicing in vitro, and splicing can be restored by the addition of RNAs containing an hnRNP A1 binding site to the reaction, demonstrating that hnRNP A1 inhibits splicing of the late HPV-16 mRNAs through the splicing silencer sequence. While we show that one role of the splicing silencer is to determine the ratio between partially spliced L2/L1 mRNAs and spliced L1 mRNAs, we also demonstrate that it inhibits splicing from the major 5' splice site in the early region to the L1 3' splice site, thereby playing an essential role in preventing late gene expression at an early stage of the viral life cycle. We speculate that the activity of the splicing silencer and possibly the concentration of hnRNP A1 in the HPV-16-infected cell determines the ability of the virus to establish a persistent infection which remains undetected by the host immune surveillance.
Figures




Similar articles
-
Identification of a 17-nucleotide splicing enhancer in HPV-16 L1 that counteracts the effect of multiple hnRNP A1-binding splicing silencers.Virology. 2007 Dec 20;369(2):351-63. doi: 10.1016/j.virol.2007.08.002. Epub 2007 Sep 14. Virology. 2007. PMID: 17869320
-
A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression.J Virol. 2005 Sep;79(18):12002-15. doi: 10.1128/JVI.79.18.12002-12015.2005. J Virol. 2005. PMID: 16140776 Free PMC article.
-
A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein.J Virol. 2005 Apr;79(7):4270-88. doi: 10.1128/JVI.79.7.4270-4288.2005. J Virol. 2005. PMID: 15767428 Free PMC article.
-
HPV-16 RNA processing.Front Biosci. 2008 May 1;13:5880-91. doi: 10.2741/3123. Front Biosci. 2008. PMID: 18508629 Review.
-
Papillomavirus transcripts and posttranscriptional regulation.Virology. 2013 Oct;445(1-2):187-96. doi: 10.1016/j.virol.2013.04.034. Epub 2013 May 23. Virology. 2013. PMID: 23706315 Review.
Cited by
-
Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer.Am J Hum Genet. 2007 Mar;80(3):416-32. doi: 10.1086/511992. Epub 2007 Jan 18. Am J Hum Genet. 2007. PMID: 17273963 Free PMC article.
-
Suppression of HPV-16 late L1 5'-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs.Nucleic Acids Res. 2013 Dec;41(22):10488-508. doi: 10.1093/nar/gkt803. Epub 2013 Sep 5. Nucleic Acids Res. 2013. PMID: 24013563 Free PMC article.
-
Splicing and Polyadenylation of Human Papillomavirus Type 16 mRNAs.Int J Mol Sci. 2017 Feb 9;18(2):366. doi: 10.3390/ijms18020366. Int J Mol Sci. 2017. PMID: 28208770 Free PMC article. Review.
-
Papillomavirus genome structure, expression, and post-transcriptional regulation.Front Biosci. 2006 Sep 1;11:2286-302. doi: 10.2741/1971. Front Biosci. 2006. PMID: 16720315 Free PMC article. Review.
-
The E1circumflexE4 protein of human papillomavirus interacts with the serine-arginine-specific protein kinase SRPK1.J Virol. 2007 Jun;81(11):5437-48. doi: 10.1128/JVI.02609-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360743 Free PMC article.
References
-
- Baker, C. C. 1997. Posttranscriptional regulation of papillomavirus gene expression, p. 11-16. In S. R. Billakanti, C. E. Calef, A. D. Farmer, A. L. Halpern, and G. L. Myres (ed.), Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

