Splice junction map of simian parvovirus transcripts
- PMID: 15452211
- PMCID: PMC521819
- DOI: 10.1128/JVI.78.20.10911-10919.2004
Splice junction map of simian parvovirus transcripts
Abstract
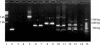
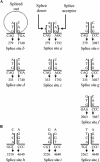
The transcription map of simian parvovirus (SPV), an Erythrovirus similar to Parvovirus B19, was investigated. RNA was extracted from tissues of experimentally infected cynomolgus macaques and subjected to reverse transcription-PCR with SPV-specific primers. The PCR products were cloned and sequenced to identify splice junctions. A total of 14 distinct sequences were identified as putative partial transcripts. Of these, 13 were spliced; a single unspliced transcript putatively encoded NS1. Sequence analysis revealed that spliced partial transcripts may encode portions of open reading frames for the major capsid proteins VP1 and VP2 and smaller, unknown proteins. These unspliced and spliced transcripts and putative proteins encoded by SPV were similar to those of B19. Initial splice junctions at nucleotides 279 and 333 were analogous to those at nucleotides 406 and 441, respectively, in B19. Seven of the 10 splices identified had typical GT/AG donor/acceptor junctions. The splice sites were confirmed by Northern blotting and autoradiography. In contrast to B19, which has a maximum of two splices per transcript, up to three splices were observed in SPV transcripts. A spliced transcript putatively encoding a truncated version of NS1, as seen with minute virus of mice and adeno-associated virus 2, was also observed. The findings indicate that that the splicing pattern of transcripts of SPV and B19 is similar, but SPV also has coding strategies in common with other parvoviruses.
Figures



References
-
- Beard, C., J. St Amand, and C. R. Astell. 1989. Transient expression of B19 parvovirus gene products in COS-7 cells transfected with B19-SV40 hybrid vectors. Virology 172:659-664. - PubMed
-
- Berns, K. I., M. Bergoin, M. Bloom, M. Lederman, N. Muzyczka, G. Siegl, J. Tal, and P. Tattersall. 2000. Family Parvoviridae, p. 311-323. In M. H. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses: seventh report of the International Committee on Taxonomy of Viruses. Virology Division, International Union of Microbiological Societies, San Diego, Calif.
-
- Blundell, M. C., C. Beard, and C. R. Astell. 1987. In vitro identification of a B19 parvovirus promoter. Virology 157:534-538. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

