mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers
- PMID: 15452213
- PMCID: PMC521861
- DOI: 10.1128/JVI.78.20.10927-10938.2004
mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers
Abstract
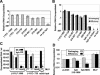
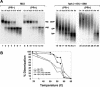
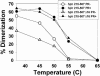
Prior work by others has shown that insertion of psi (i.e., leader) sequences from the Moloney murine leukemia virus (MLV) genome into the 3' untranslated region of a nonviral mRNA leads to the specific encapsidation of this RNA in MLV particles. We now report that these RNAs are, like genomic RNAs, encapsidated as dimers. These dimers have the same thermostability as MLV genomic RNA dimers; like them, these dimers are more stable if isolated from mature virions than from immature virions. We characterized encapsidated mRNAs containing deletions or truncations of MLV psi or with psi sequences from MLV-related acute transforming viruses. The results indicate that the dimeric linkage in genomic RNA can be completely attributed to the psi region of the genome. While this conclusion agrees with earlier electron microscopic studies on mature MLV dimers, it is the first evidence as to the site of the linkage in immature dimers for any retrovirus. Since the Psi(+) mRNA is not encapsidated as well as genomic RNA, it is only present in a minority of virions. The fact that it is nevertheless dimeric argues strongly that two of these molecules are packaged into particles together. We also found that the kissing loop is unnecessary for this coencapsidation or for the stability of mature dimers but makes a major contribution to the stability of immature dimers. Our results are consistent with the hypothesis that the packaging signal involves a dimeric structure in which the RNAs are joined by intermolecular interactions between GACG loops.
Figures


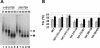


Similar articles
-
Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Psi-RNA packaging signal: implications for genome recognition.J Mol Biol. 2001 Nov 23;314(2):217-32. doi: 10.1006/jmbi.2001.5139. J Mol Biol. 2001. PMID: 11718556
-
Preliminary physical mapping of RNA-RNA linkages in the genomic RNA of Moloney murine leukemia virus.J Virol. 2005 Jul;79(13):8142-8. doi: 10.1128/JVI.79.13.8142-8148.2005. J Virol. 2005. PMID: 15956559 Free PMC article.
-
Analytical study of rat retrotransposon VL30 RNA dimerization in vitro and packaging in murine leukemia virus.J Mol Biol. 1994 Jul 29;240(5):434-44. doi: 10.1006/jmbi.1994.1459. J Mol Biol. 1994. PMID: 8046749
-
Retroviral RNA packaging: a review.Arch Virol Suppl. 1994;9:513-22. doi: 10.1007/978-3-7091-9326-6_49. Arch Virol Suppl. 1994. PMID: 8032280 Review.
-
From Cells to Virus Particles: Quantitative Methods to Monitor RNA Packaging.Viruses. 2016 Aug 22;8(8):239. doi: 10.3390/v8080239. Viruses. 2016. PMID: 27556480 Free PMC article. Review.
Cited by
-
Cross- and Co-Packaging of Retroviral RNAs and Their Consequences.Viruses. 2016 Oct 11;8(10):276. doi: 10.3390/v8100276. Viruses. 2016. PMID: 27727192 Free PMC article. Review.
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
-
Bipartite viral RNA genome heterodimerization influences genome packaging and virion thermostability.J Virol. 2024 Mar 19;98(3):e0182023. doi: 10.1128/jvi.01820-23. Epub 2024 Feb 8. J Virol. 2024. PMID: 38329331 Free PMC article.
-
Structural dynamics of retroviral genome and the packaging.Front Microbiol. 2011 Dec 29;2:264. doi: 10.3389/fmicb.2011.00264. eCollection 2011. Front Microbiol. 2011. PMID: 22232618 Free PMC article.
-
Murine leukemia virus RNA dimerization is coupled to transcription and splicing processes.Retrovirology. 2010 Aug 5;7:64. doi: 10.1186/1742-4690-7-64. Retrovirology. 2010. PMID: 20687923 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

