Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response
- PMID: 15452220
- PMCID: PMC521853
- DOI: 10.1128/JVI.78.20.10995-11006.2004
Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response
Abstract
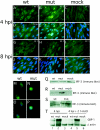
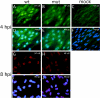
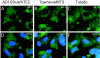
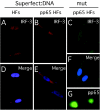
We have identified a cytomegalovirus virion protein capable of modulating the rapid induction of an interferon-like response in cells that follows virus binding and penetration. Functional genomics revealed a role for the major cytomegalovirus structural protein, pp65 (ppUL83), in counteracting this response. The underlying mechanism involves a differential impact of this structural protein on the regulation of interferon response factor 3 (IRF-3). In contrast, NF-kappaB is activated independent of pp65, and neither STAT1 nor STAT3 becomes activated by either virus. pp65 is sufficient to prevent the activation of IRF-3 when introduced alone into cells. pp65 acts by inhibiting nuclear accumulation of IRF-3 and is associated with a reduced IRF-3 phosphorylation state. Thus, this investigation shows that the major structural protein of cytomegalovirus is committed to the modulation of the IRF-3 response, a primary mediator of the type I interferon response. By subverting IRF-3, the virus escapes throwing a central alarm devoted to both immediate antiviral control and regulation of the immune response.
Figures



References
-
- Arrode, G., and C. Davrinche. 2003. Dendritic cells and HCMV cross-presentation. Curr. Top. Microbiol. Immunol. 276:277-294. - PubMed
-
- Au, W. C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. - PMC - PubMed
-
- Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

