APOBEC3G genetic variants and their influence on the progression to AIDS
- PMID: 15452227
- PMCID: PMC521814
- DOI: 10.1128/JVI.78.20.11070-11076.2004
APOBEC3G genetic variants and their influence on the progression to AIDS
Abstract
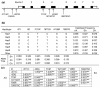
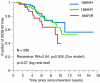
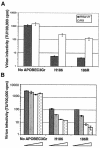
The cytosine deaminase APOBEC3G, in the absence of the human immunodeficiency virus type 1 (HIV-1) accessory gene HIV-1 viral infectivity factor (vif), inhibits viral replication by introducing G-->A hypermutation in the newly synthesized HIV-1 DNA negative strand. We tested the hypothesis that genetic variants of APOBEC3G may modify HIV-1 transmission and disease progression. Single nucleotide polymorphisms were identified in the promoter region (three), introns (two), and exons (two). Genotypes were determined for 3,073 study participants enrolled in six HIV-AIDS prospective cohorts. One codon-changing variant, H186R in exon 4, was polymorphic in African Americans (AA) (f = 37%) and rare in European Americans (f < 3%) or Europeans (f = 5%). For AA, the variant allele 186R was strongly associated with decline in CD4 T cells (CD4 slope on square root scale: -1.86, P = 0.009), The 186R allele was also associated with accelerated progression to AIDS-defining conditions in AA. The in vitro antiviral activity of the 186R enzyme was not inferior to that of the common H186 variant. These studies suggest that there may be a modifying role of variants of APOBEC3G on HIV-1 disease progression that warrants further investigation.
Figures
Similar articles
-
APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load.AIDS. 2010 Jan 16;24(2):195-204. doi: 10.1097/QAD.0b013e3283353bba. AIDS. 2010. PMID: 19996938 Free PMC article.
-
Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation.J Virol. 2006 Sep;80(18):9259-69. doi: 10.1128/JVI.00888-06. J Virol. 2006. PMID: 16940537 Free PMC article.
-
APOBEC3G genetic variants and their association with risk of HIV infection in highly exposed Caucasians.AIDS. 2006 Oct 3;20(15):1984-6. doi: 10.1097/01.aids.0000247124.35129.e1. AIDS. 2006. PMID: 16988524
-
Protecting APOBEC3G: a potential new target for HIV drug discovery.Curr Opin Investig Drugs. 2005 Feb;6(2):141-7. Curr Opin Investig Drugs. 2005. PMID: 15751736 Review.
-
New insights into the role of Vif in HIV-1 replication.AIDS Rev. 2004 Jan-Mar;6(1):34-9. AIDS Rev. 2004. PMID: 15168739 Review.
Cited by
-
Downregulation of APOBEC3G by xenotropic murine leukemia-virus related virus (XMRV) in prostate cancer cells.Virol J. 2011 Dec 12;8:531. doi: 10.1186/1743-422X-8-531. Virol J. 2011. PMID: 22152111 Free PMC article.
-
Host Factors and HIV-1 Replication: Clinical Evidence and Potential Therapeutic Approaches.Front Immunol. 2013 Oct 24;4:343. doi: 10.3389/fimmu.2013.00343. Front Immunol. 2013. PMID: 24167505 Free PMC article. Review.
-
HIV-1 and human genetic variation.Nat Rev Genet. 2021 Oct;22(10):645-657. doi: 10.1038/s41576-021-00378-0. Epub 2021 Jun 24. Nat Rev Genet. 2021. PMID: 34168330 Free PMC article. Review.
-
Functional characterization of Vif proteins from HIV-1 infected patients with different APOBEC3G haplotypes.AIDS. 2016 Jul 17;30(11):1723-9. doi: 10.1097/QAD.0000000000001113. AIDS. 2016. PMID: 27064995 Free PMC article.
-
The role of natural killer (NK) cells and NK cell receptor polymorphisms in the assessment of HIV-1 neutralization.PLoS One. 2012;7(4):e29454. doi: 10.1371/journal.pone.0029454. Epub 2012 Apr 11. PLoS One. 2012. PMID: 22509241 Free PMC article.
References
-
- An, P., D. Vlahov, J. B. Margolick, J. Phair, T. R. O'Brien, J. Lautenberger, S. J. O'Brien, and C. A. Winkler. 2003. A tumor necrosis factor-alpha-inducible promoter variant of interferon-gamma accelerates CD4+ T cell depletion in human immunodeficiency virus-1-infected individuals. J. Infect. Dis. 188:228-231. - PubMed
-
- Bleiber, G., M. May, C. Suarez, R. Martinez, C. Marzolini, M. Egger, and A. Telenti. 2004. MDR1 genetic polymorphism does not modify either cell permissiveness to HIV-1 or disease progression before treatment. J. Infect. Dis. 189:583-586. - PubMed
-
- Buchbinder, S. P., M. H. Katz, N. A. Hessol, P. M. O'Malley, and S. D. Holmberg. 1994. Long-term HIV-1 infection without immunologic progression. AIDS 8:1123-1128. - PubMed
-
- Celentano, D. D., N. Galai, A. K. Sethi, N. G. Shah, S. A. Strathdee, D. Vlahov, and J. E. Gallant. 2001. Time to initiating highly active antiretroviral therapy among HIV-infected injection drug users. AIDS 15:1707-1715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

