Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3
- PMID: 15452230
- PMCID: PMC521805
- DOI: 10.1128/JVI.78.20.11097-11107.2004
Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3
Abstract
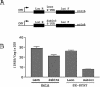
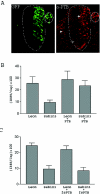
Mutations critical for the central nervous system (CNS) attenuation of the Sabin vaccine strains of poliovirus (PV) are located within the viral internal ribosome entry site (IRES). We examined the interaction of the IRESs of PV type 3 (PV3) and Sabin type 3 (Sabin3) with polypyrimidine tract-binding protein (PTB) and a neural cell-specific homologue, nPTB. PTB and nPTB were found to bind to a site directly adjacent to the attenuating mutation, and binding at this site was less efficient on the Sabin3 IRES than on the PV3 IRES. Translation mediated by the PV3 and Sabin3 IRESs in neurons of the chicken embryo spinal cord demonstrated a translation deficit for the Sabin3 IRES that could be rescued by increasing PTB expression in the CNS. These data suggest that the low levels of PTB available in the CNS, coupled to a reduced binding of PTB on the Sabin3 IRES, leads to its CNS-specific attenuation. This study also demonstrates the use of the chicken embryo to easily investigate translation of RNA within a neuron in the CNS of an intact living organism.
Figures




References
-
- Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

