Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1
- PMID: 15452240
- PMCID: PMC521858
- DOI: 10.1128/JVI.78.20.11208-11218.2004
Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1
Abstract
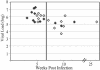
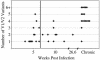
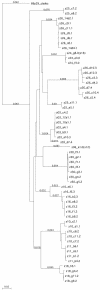
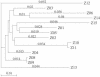
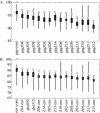
Human immunodeficiency virus type 1 (HIV-1) exists as a complex population of multiple genotypic variants in persons with chronic infection. However, acute HIV-1 infection via sexual transmission is a low-probability event in which there is thought to be low genetic complexity in the initial inoculum. In order to assess the viral complexity present during primary HIV-1 infection, the V1/V2 and V3 variable regions of the env gene were examined by using a heteroduplex tracking assay (HTA) capable of resolving these genotypic variants. Blood plasma samples from 26 primary HIV-1-infected subjects were analyzed for their level of diversity. Half of the subjects had more than one V1/V2 viral variant during primary infection, indicating the frequent transmission of multiple variants. This observation is inconsistent with the idea of infrequent transmission based on a small transmitting inoculum of cell-free virus. In chronically infected subjects, the complexity of the viral populations was even greater in both the V1/V2 and the V3 regions than in acutely infected subjects, indicating that in spite of the presence of multiple variants in acute infection, the virus does pass through a genetic bottleneck during transmission. We also examined how well the infecting virus penetrated different anatomical compartments by using the HTA. Viral variants detected in blood plasma were compared to those detected in seminal plasma and/or cerebral spinal fluid of six individuals. The virus in each of these compartments was to a large extent identical to virus in blood plasma, a finding consistent with rapid penetration of the infecting variant(s). The low-probability transmission of multiple variants could be the result of transient periods of hyperinfectiousness or hypersusceptibility. Alternatively, the inefficient transfer of a multiply infected cell could account for both the low probability of transmission and the transfer of multiple variants.
Figures
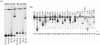



References
-
- Betts, M. R., J. Krowka, C. Santamaria, K. Balsamo, F. Gao, G. Mulundu, C. Luo, N. N′Gandu, H. Sheppard, B. H. Hahn, S. Allen, and J. A. Frelinger. 1997. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J. Virol. 71:8908-8911. - PMC - PubMed
-
- Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. - PubMed
-
- Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. - PubMed
-
- Chakraborty, H., P. K. Sen, R. W. Helms, P. L. Vernazza, S. A. Fiscus, J. J. Eron, B. K. Patterson, R. W. Coombs, J. N. Krieger, and M. S. Cohen. 2001. Viral burden in genital secretions determines male-to-female sexual transmission of HIV-1: a probabilistic empiric model. AIDS 15:621-627. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- T32 AI 007419/AI/NIAID NIH HHS/United States
- M01 RR000046/RR/NCRR NIH HHS/United States
- K23 AI 01781/AI/NIAID NIH HHS/United States
- T32 AI007419/AI/NIAID NIH HHS/United States
- RR 00046/RR/NCRR NIH HHS/United States
- K24 AI001608/AI/NIAID NIH HHS/United States
- R01 DK049381/DK/NIDDK NIH HHS/United States
- K23 AI001781/AI/NIAID NIH HHS/United States
- R01 DK 49381/DK/NIDDK NIH HHS/United States
- R01 AI044667/AI/NIAID NIH HHS/United States
- P30 AI 50410/AI/NIAID NIH HHS/United States
- R01 AI 44667/AI/NIAID NIH HHS/United States
- K24 AI 01608/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

