A yeast arginine specific tRNA is a remnant aspartate acceptor
- PMID: 15452274
- PMCID: PMC521656
- DOI: 10.1093/nar/gkh843
A yeast arginine specific tRNA is a remnant aspartate acceptor
Abstract
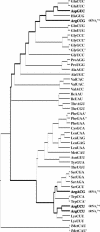
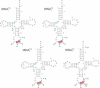
High specificity in aminoacylation of transfer RNAs (tRNAs) with the help of their cognate aminoacyl-tRNA synthetases (aaRSs) is a guarantee for accurate genetic translation. Structural and mechanistic peculiarities between the different tRNA/aaRS couples, suggest that aminoacylation systems are unrelated. However, occurrence of tRNA mischarging by non-cognate aaRSs reflects the relationship between such systems. In Saccharomyces cerevisiae, functional links between arginylation and aspartylation systems have been reported. In particular, it was found that an in vitro transcribed tRNAAsp is a very efficient substrate for ArgRS. In this study, the relationship of arginine and aspartate systems is further explored, based on the discovery of a fourth isoacceptor in the yeast genome, tRNA4Arg. This tRNA has a sequence strikingly similar to that of tRNAAsp but distinct from those of the other three arginine isoacceptors. After transplantation of the full set of aspartate identity elements into the four arginine isoacceptors, tRNA4Arg gains the highest aspartylation efficiency. Moreover, it is possible to convert tRNA4Arg into an aspartate acceptor, as efficient as tRNAAsp, by only two point mutations, C38 and G73, despite the absence of the major anticodon aspartate identity elements. Thus, cryptic aspartate identity elements are embedded within tRNA4Arg. The latent aspartate acceptor capacity in a contemporary tRNAArg leads to the proposal of an evolutionary link between tRNA4Arg and tRNAAsp genes.
Figures


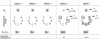

References
-
- de Duve C. (1988) The second genetic code. Nature, 333, 117–118. - PubMed
-
- Kisselev L. (1985) The role of the anticodon in recognition of tRNA by aminoacyl-tRNA synthetases. Prog. Nucleic Acid Res. Mol. Biol., 32, 237–266. - PubMed
-
- Schulman L.H. and Pelka,H. (1988) Anticodon switching changes the identity of methionine and valine transfer RNAs. Science, 242, 765–768. - PubMed
-
- Hou Y.-M. and Schimmel,P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature, 333, 140–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

