A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation
- PMID: 15452344
- PMCID: PMC521940
- DOI: 10.1073/pnas.0405786101
A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation
Abstract
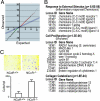
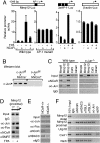
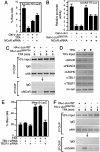
The nuclear receptor corepressor (NCoR) and the related factor known as silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) are essential components of multiprotein complexes that mediate active repression by unliganded nuclear receptors. Recent studies suggest that NCoR and SMRT can interact with and exert repressive effects on several other classes of DNA-binding transcription factors, but the physiological importance of these interactions has not been established. Here, investigation of endogenous transcriptional programs regulated by NCoR in macrophages reveals that NCoR acts as a transcriptional checkpoint for activator protein (AP)-1-dependent gene networks that regulate diverse biological processes including inflammation, cell migration, and collagen catabolism, with loss of NCoR, resulting in derepression of AP-1 target genes. The NCoR corepressor complex imposes an active block of exchange of c-Jun for c-Jun/c-Fos heterodimers, with targeted deletion of the c-Jun locus, resulting in loss of NCoR complexes from AP-1 target genes under basal conditions. The checkpoint function of NCoR is relieved by signal-dependent phosphorylation of c-Jun, which directs removal of NCoR/HDAC3/TBL1/TBLR1 complexes through recruitment of a specific ubiquitylation complex, as a prerequisite to the default binding of c-Jun/c-Fos heterodimers and transcriptional activation. The requirement for a checkpoint function to achieve the appropriate dynamic range of transcriptional responses to inflammatory signals is likely to be used by other signal-dependent transcription factors that regulate diverse homeostatic and developmental processes.
Figures


References
-
- Rosenfeld, M. G. & Glass, C. K. (2001) J. Biol. Chem. 276, 36865-36868. - PubMed
-
- Naar, A. M., Lemon, B. D. & Tjian, R. (2001) Annu. Rev. Biochem. 70, 475-501. - PubMed
-
- McKenna, N. J. & O'Malley, B. W. (2002) Cell 108, 465-474. - PubMed
-
- Horlein, A. J., Naar, A. M., Heinzel, T., Torchia, J., Gloss, B., Kurokawa, R., Ryan, A., Kamei, Y., Soderstrom, M., Glass, C. K., et al. (1995) Nature 377, 397-404. - PubMed
-
- Chen, J. D. & Evans, R. M. (1995) Nature 377, 454-457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

