A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions
- PMID: 15453917
- PMCID: PMC523849
- DOI: 10.1186/1471-2105-5-139
A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions
Abstract
Background: MicroRNAs are approximately 17-24 nt. noncoding RNAs found in all eukaryotes that degrade messenger RNAs via RNA interference (if they bind in a perfect or near-perfect complementarity to the target mRNA), or arrest translation (if the binding is imperfect). Several microRNA targets have been identified in lower organisms, but only one mammalian microRNA target has yet been validated experimentally.
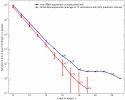
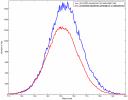
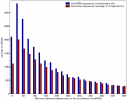
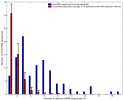
Results: We carried out a population-wide statistical analysis of how human microRNAs interact complementarily with human mRNAs, looking for characteristics that differ significantly as compared with scrambled control sequences. These characteristics were used to identify a set of 71 outlier mRNAs unlikely to have been hit by chance. Unlike the case in C. elegans and Drosophila, many human microRNAs exhibited long exact matches (10 or more bases in a row), up to and including perfect target complementarity. Human microRNAs hit outlier mRNAs within the protein coding region about 2/3 of the time. And, the stretches of perfect complementarity within microRNA hits onto outlier mRNAs were not biased near the 5'-end of the microRNA. In several cases, an individual microRNA hit multiple mRNAs that belonged to the same functional class.
Conclusions: The analysis supports the notion that sequence complementarity is the basis by which microRNAs recognize their biological targets, but raises the possibility that human microRNA-mRNA target interactions follow different rules than have been previously characterized in Drosophila and C. elegans.
Figures
References
-
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

