Sculpting the proteome with AAA(+) proteases and disassembly machines
- PMID: 15454077
- PMCID: PMC2717008
- DOI: 10.1016/j.cell.2004.09.020
Sculpting the proteome with AAA(+) proteases and disassembly machines
Abstract
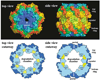
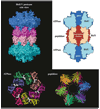
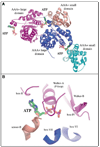
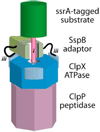
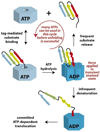
Machines of protein destruction-including energy-dependent proteases and disassembly chaperones of the AAA(+) ATPase family-function in all kingdoms of life to sculpt the cellular proteome, ensuring that unnecessary and dangerous proteins are eliminated and biological responses to environmental change are rapidly and properly regulated. Exciting progress has been made in understanding how AAA(+) machines recognize specific proteins as targets and then carry out ATP-dependent dismantling of the tertiary and/or quaternary structure of these molecules during the processes of protein degradation and the disassembly of macromolecular complexes.
Figures
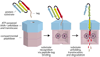
References
-
- Beuron F, Maurizi MR, Belnap DM, Kocsis E, Booy FP, Kessel M, Steven AC. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J. Struct. Biol. 1998;123:248–259. - PubMed
-
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. - PubMed
-
- Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent tethering of the SspB adaptor to the AAA+ protease ClpXP is required for efficient substrate delivery: a protein-design study. Mol. Cell. 2004a;13:443–449. - PubMed
-
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004b in press. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

