A bending mode analysis for growing microtubules: evidence for a velocity-dependent rigidity
- PMID: 15454464
- PMCID: PMC1304691
- DOI: 10.1529/biophysj.103.038877
A bending mode analysis for growing microtubules: evidence for a velocity-dependent rigidity
Abstract
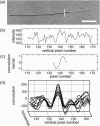
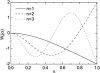
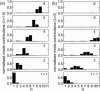
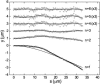
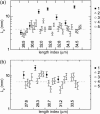
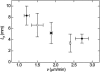
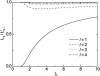
Microtubules are dynamic protein polymers that continuously switch between elongation and rapid shrinkage. They have an exceptional bending stiffness that contributes significantly to the mechanical properties of eukaryotic cells. Measurements of the persistence length of microtubules have been published since 10 years but the reported values vary over an order of magnitude without an available explanation. To precisely measure the rigidity of microtubules in their native growing state, we adapted a previously developed bending mode analysis of thermally driven shape fluctuations to the case of an elongating filament that is clamped at one end. Microtubule shapes were quantified using automated image processing, allowing for the characterization of up to five bending modes. When taken together with three other less precise measurements, our rigidity data suggest that fast-growing microtubules are less stiff than slow-growing microtubules. This would imply that care should be taken in interpreting rigidity measurements on stabilized microtubules whose growth history is not known. In addition, time analysis of bending modes showed that higher order modes relax more slowly than expected from simple hydrodynamics, possibly by the effects of internal friction within the microtubule.
Copyright 2004 Biophysical Society
Figures




References
-
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular Biology of the Cell, 4th ed. Garland Science, New York.
-
- Cassimeris, L., D. Gard, P. T. Tran, and H. P. Erickson. 2001. XMAP215 is a long thin molecule that does not increase microtubule stiffness. J. Cell Sci. 114:3025–3033. - PubMed
-
- Chretien, D., and S. D. Fuller. 2000. Microtubules switch occasionally into unfavorable configurations during elongation. J. Mol. Biol. 298:663–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

