Abortive transposition by a group II intron in yeast mitochondria
- PMID: 15454528
- PMCID: PMC1448100
- DOI: 10.1534/genetics.104.027003
Abortive transposition by a group II intron in yeast mitochondria
Abstract
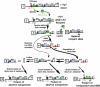
Group II intron homing in yeast mitochondria is initiated at active target sites by activities of intron-encoded ribonucleoprotein (RNP) particles, but is completed by competing recombination and repair mechanisms. Intron aI1 transposes in haploid cells at low frequency to target sites in mtDNA that resemble the exon 1-exon 2 (E1/E2) homing site. This study investigates a system in which aI1 can transpose in crosses (i.e., in trans). Surprisingly, replacing an inefficient transposition site with an active E1/E2 site supports <1% transposition of aI1. Instead, the ectopic site was mainly converted to the related sequence in donor mtDNA in a process we call "abortive transposition." Efficient abortive events depend on sequences in both E1 and E2, suggesting that most events result from cleavage of the target site by the intron RNP particles, gapping, and recombinational repair using homologous sequences in donor mtDNA. A donor strain that lacks RT activity carries out little abortive transposition, indicating that cDNA synthesis actually promotes abortive events. We also infer that some intermediates abort by ejecting the intron RNA from the DNA target by forward splicing. These experiments provide new insights to group II intron transposition and homing mechanisms in yeast mitochondria.
Figures




References
-
- Adams, A., D. E. Gottschling, C. A. Kaiser and T. Stearns, 1997 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Aizawa, Y., Q. Xiang, A. M. Lambowitz and A. M. Pyle, 2003. The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol. Cell 11: 795–805. - PubMed
-
- Belfort, M., V. Derbyshire, M. M. Parker, B. Cousineau and A. M. Lambowitz, 2002 Mobile introns: pathways and proteins, pp. 761–783 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society for Microbiology, Washington, DC.
-
- Blanc, H., B. Dujon, M. Guerineau and P. P. Slonimski, 1978. Detection of specific DNA sequences in yeast by colony hybridization. Mol. Gen. Genet. 161: 311–315. - PubMed
-
- Butow, R. A., R. M. Henke, J. V. Moran, S. C. Belcher and P. S. Perlman, 1996 Transformation of Saccharomyces cerevisiae mitochondria using the biolistic gun, pp. 265–278 in Methods in Enzymology, edited by G. Attardi and A. Chomyn. Academic Press, San Diego/New York/Boston. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

