A genetic screen for dominant modifiers of a cyclin E hypomorphic mutation identifies novel regulators of S-phase entry in Drosophila
- PMID: 15454540
- PMCID: PMC1448096
- DOI: 10.1534/genetics.104.026617
A genetic screen for dominant modifiers of a cyclin E hypomorphic mutation identifies novel regulators of S-phase entry in Drosophila
Abstract
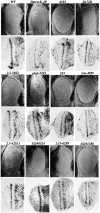
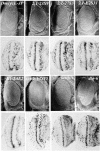
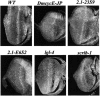
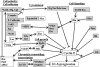
Cyclin E together with its kinase partner Cdk2 is a critical regulator of entry into S phase. To identify novel genes that regulate the G1- to S-phase transition within a whole animal we made use of a hypomorphic cyclin E mutation, DmcycEJP, which results in a rough eye phenotype. We screened the X and third chromosome deficiencies, tested candidate genes, and carried out a genetic screen of 55,000 EMS or X-ray-mutagenized flies for second or third chromosome mutations that dominantly modified the DmcycEJP rough eye phenotype. We have focused on the DmcycEJP suppressors, S(DmcycEJP), to identify novel negative regulators of S-phase entry. There are 18 suppressor gene groups with more than one allele and several genes that are represented by only a single allele. All S(DmcycEJP) tested suppress the DmcycEJP rough eye phenotype by increasing the number of S phases in the postmorphogenetic furrow S-phase band. By testing candidates we have identified several modifier genes from the mutagenic screen as well as from the deficiency screen. DmcycEJP suppressor genes fall into the classes of: (1) chromatin remodeling or transcription factors; (2) signaling pathways; and (3) cytoskeletal, (4) cell adhesion, and (5) cytoarchitectural tumor suppressors. The cytoarchitectural tumor suppressors include scribble, lethal-2-giant-larvae (lgl), and discs-large (dlg), loss of function of which leads to neoplastic tumors and disruption of apical-basal cell polarity. We further explored the genetic interactions of scribble with S(DmcycEJP) genes and show that hypomorphic scribble mutants exhibit genetic interactions with lgl, scab (alphaPS3-integrin--cell adhesion), phyllopod (signaling), dEB1 (microtubule-binding protein--cytoskeletal), and moira (chromatin remodeling). These interactions of the cytoarchitectural suppressor gene, scribble, with cell adhesion, signaling, cytoskeletal, and chromatin remodeling genes, suggest that these genes may act in a common pathway to negatively regulate cyclin E or S-phase entry.
Figures
References
-
- Adler, P. N., and H. Lee, 2001. Frizzled signaling and cell-cell interactions in planar polarity. Curr. Opin. Cell Biol. 13: 635–640. - PubMed
-
- Ahmed, Y., S. Hayashi, A. Levine and E. Wieschaus, 1998. Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell 93: 1171–1182. - PubMed
-
- Allenspach, E. J., I. Maillard, J. C. Aster and W. S. Pear, 2002. Notch signaling in cancer. Cancer Biol. Ther. 1: 466–476. - PubMed
-
- Amaldi, F., and P. Pierandrei-Amaldi, 1997. TOP genes: a translationally controlled class of genes including those coding for ribosomal proteins. Prog. Mol. Subcell. Biol. 18: 1–17. - PubMed
-
- Araujo, H., E. Negreiros and E. Bier, 2003. Integrins modulate Sog activity in the Drosophila wing. Development 130: 3851–3864. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

