Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells
- PMID: 15456859
- PMCID: PMC517870
- DOI: 10.1128/MCB.24.20.8834-8846.2004
Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells
Abstract
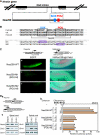
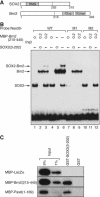
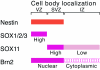
Intermediate-filament Nestin and group B1 SOX transcription factors (SOX1/2/3) are often employed as markers for neural primordium, suggesting their regulatory link. We have identified adjacent and essential SOX and POU factor binding sites in the Nestin neural enhancer. The 30-bp sequence of the enhancer including these sites (Nes30) showed a nervous system-specific and SOX-POU-dependent enhancer activity in multimeric forms in transfection assays and was utilized in assessing the specificity of the synergism; combinations of either group B1 or group C SOX (SOX11) with class III POU proved effective. In embryonic day 13.5 mouse spinal cord, Nestin was expressed in the cells with nuclei in the ventricular and subventricular zones. SOX1/2/3 expression was confined to the nuclei of the ventricular zone; SOX11 localized to the nuclei of both subventricular (high-level expression) and intermediate (low-level expression) zones. Class III POU (Brn2) was expressed at high levels, localizing to the nucleus in the ventricular and subventricular zones; moderate expression was observed in the intermediate zone, distributed in the cytoplasm. These data support the model that synergic interactions between group B1/C SOX and class III POU within the nucleus determine Nestin expression. Evidence also suggests that such interactions are involved in the regulation of neural primordial cells.
Figures






References
-
- Alvarez-Bolado, G., M. G. Rosenfeld, and L. W. Swanson. 1995. Model of forebrain regionalization based on spatiotemporal patterns of POU-III homeobox gene expression, birthdates, and morphological features. J. Comp. Neurol. 355:237-295. - PubMed
-
- Alvarez-Buylla, A., B. Seri, and F. Doetsch. 2002. Identification of neural stem cells in the adult vertebrate brain. Brain Res. Bull. 57:751-758. - PubMed
-
- Andersen, B., and M. G. Rosenfeld. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 22:2-35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
