Bone-specific transcription factor Runx2 interacts with the 1alpha,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells
- PMID: 15456860
- PMCID: PMC517904
- DOI: 10.1128/MCB.24.20.8847-8861.2004
Bone-specific transcription factor Runx2 interacts with the 1alpha,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells
Abstract
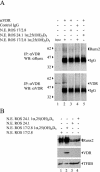
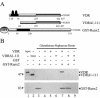
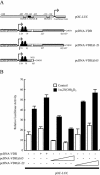
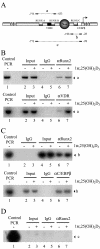
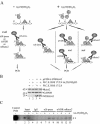
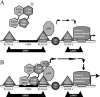
Bone-specific transcription of the osteocalcin (OC) gene is regulated principally by the Runx2 transcription factor and is further stimulated in response to 1alpha,25-dihydroxyvitamin D3 via its specific receptor (VDR). The rat OC gene promoter contains three recognition sites for Runx2 (sites A, B, and C). Mutation of sites A and B, which flank the 1alpha,25-dihydroxyvitamin D3-responsive element (VDRE), abolishes 1alpha,25-dihydroxyvitamin D3-dependent enhancement of OC transcription, indicating a tight functional relationship between the VDR and Runx2 factors. In contrast to most of the members of the nuclear receptor family, VDR possesses a very short N-terminal A/B domain, which has led to the suggestion that its N-terminal region does not contribute to transcriptional enhancement. Here, we have combined transient-overexpression, coimmunoprecipitation, in situ colocalization, chromatin immunoprecipitation, and glutathione S-transferase pull-down analyses to demonstrate that in osteoblastic cells expressing OC, VDR interacts directly with Runx2 bound to site B, which is located immediately adjacent to the VDRE. This interaction contributes significantly to 1alpha,25-dihydroxyvitamin D3-dependent enhancement of the OC promoter and requires a region located C terminal to the runt homology DNA binding domain of Runx2 and the N-terminal region of VDR. Together, our results indicate that Runx2 plays a key role in the 1alpha,25-dihydroxyvitamin D3-dependent stimulation of the OC promoter in osteoblastic cells by further stabilizing the interaction of the VDR with the VDRE. These studies demonstrate a novel mechanism for combinatorial control of bone tissue-specific gene expression. This mechanism involves the intersection of two major pathways: Runx2, a "master" transcriptional regulator of osteoblast differentiation, and 1alpha,25-dihydroxyvitamin D3, a hormone that promotes expression of genes associated with these terminally differentiated bone cells.
Figures









Similar articles
-
Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity.Mol Cell Biol. 2003 May;23(9):3339-51. doi: 10.1128/MCB.23.9.3339-3351.2003. Mol Cell Biol. 2003. PMID: 12697832 Free PMC article.
-
The Runx2 transcription factor plays a key role in the 1alpha,25-dihydroxy Vitamin D3-dependent upregulation of the rat osteocalcin (OC) gene expression in osteoblastic cells.J Steroid Biochem Mol Biol. 2004 May;89-90(1-5):269-71. doi: 10.1016/j.jsbmb.2004.03.076. J Steroid Biochem Mol Biol. 2004. PMID: 15225783 Review.
-
Interaction of the 1alpha,25-dihydroxyvitamin D3 receptor at the distal promoter region of the bone-specific osteocalcin gene requires nucleosomal remodelling.Biochem J. 2002 May 1;363(Pt 3):667-76. doi: 10.1042/0264-6021:3630667. Biochem J. 2002. PMID: 11964167 Free PMC article.
-
1alpha,25-dihydroxy vitamin D3-enhanced expression of the osteocalcin gene involves increased promoter occupancy of basal transcription regulators and gradual recruitment of the 1alpha,25-dihydroxy vitamin D3 receptor-SRC-1 coactivator complex.J Cell Physiol. 2008 Mar;214(3):740-9. doi: 10.1002/jcp.21267. J Cell Physiol. 2008. PMID: 17786964
-
New understanding of the molecular mechanism of receptor-mediated genomic actions of the vitamin D hormone.Bone. 1995 Aug;17(2 Suppl):33S-38S. doi: 10.1016/8756-3282(95)00205-r. Bone. 1995. PMID: 8579895 Review.
Cited by
-
Bio-Functionalized Chitosan for Bone Tissue Engineering.Int J Mol Sci. 2021 May 31;22(11):5916. doi: 10.3390/ijms22115916. Int J Mol Sci. 2021. PMID: 34072888 Free PMC article.
-
Gene-Activated Titanium Surfaces Promote In Vitro Osteogenesis.Int J Oral Maxillofac Implants. 2017 Mar/Apr;32(2):e83–e96. doi: 10.11607/jomi.5026. Epub 2016 Oct 5. Int J Oral Maxillofac Implants. 2017. PMID: 27706263 Free PMC article.
-
Parathyroid hormone regulates osterix and Runx2 mRNA expression predominantly through protein kinase A signaling in osteoblast-like cells.J Endocrinol Invest. 2006 Feb;29(2):101-8. doi: 10.1007/BF03344081. J Endocrinol Invest. 2006. PMID: 16610234
-
p53 and MDM2 are involved in the regulation of osteocalcin gene expression.Exp Cell Res. 2012 May 1;318(8):867-76. doi: 10.1016/j.yexcr.2012.02.022. Epub 2012 Mar 3. Exp Cell Res. 2012. PMID: 22405968 Free PMC article.
-
Ezh2-dependent H3K27me3 modification dynamically regulates vitamin D3-dependent epigenetic control of CYP24A1 gene expression in osteoblastic cells.J Cell Physiol. 2020 Jun;235(6):5404-5412. doi: 10.1002/jcp.29428. Epub 2020 Jan 7. J Cell Physiol. 2020. PMID: 31907922 Free PMC article.
References
-
- Banerjee, C., L. R. McCabe, J. Y. Choi, S. W. Hiebert, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J. Cell Biochem. 66:1-8. - PubMed
-
- Christakos, S., P. Dhawan, Y. Liu, X. Peng, and A. Porta. 2003. New insights into the mechanisms of vitamin D action. J. Cell Biochem. 88:695-705. - PubMed
-
- Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. - PubMed
-
- Gutierrez, J., J. Sierra, R. Medina, M. Puchi, M. Imschenetzky, A. van Wijnen, J. Lian, G. Stein, J. Stein, and M. Montecino. 2000. Interaction of CBF alpha/AML/PEBP2 alpha transcription factors with nucleosomes containing promoter sequences requires flexibility in the translational positioning of the histone octamer and exposure of the CBF alpha site. Biochemistry 39:13565-13574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
