Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells
- PMID: 15456861
- PMCID: PMC517875
- DOI: 10.1128/MCB.24.20.8862-8871.2004
Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells
Abstract
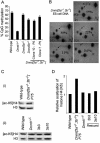
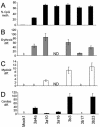
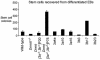
It has been reported that DNA methyltransferase 1-deficient (Dnmt1-/-) embryonic stem (ES) cells are hypomethylated (20% CpG methylation) and die through apoptosis when induced to differentiate. Here, we show that Dnmt[3a-/-,3b-/-] ES cells with just 0.6% of their CpG dinucleotides behave differently: the majority of cells within the culture are partially or completely blocked in their ability to initiate differentiation, remaining viable while retaining the stem cell characteristics of alkaline phosphatase and Oct4 expression. Restoration of DNA methylation levels rescues these defects. Severely hypomethylated Dnmt[3a-/-,3b-/-] ES cells have increased histone acetylation levels, and those cells that can differentiate aberrantly express extraembryonic markers of differentiation. Dnmt[3a-/-,3b-/-] ES cells with >10% CpG methylation are able to terminally differentiate, whereas Dnmt1-/- ES cells with 20% of the CpG methylated cannot differentiate. This demonstrates that successful terminal differentiation is not dependent simply on adequate methylation levels. There is an absolute requirement that the methylation be delivered by the maintenance enzyme Dnmt1.
Figures




References
-
- Abe, K., H. Niwa, K. Iwase, M. Takiguchi, M. Mori, S. I. Abe, and K. I. Yamamura. 1996. Endoderm-specific gene expression in embryonic stem cells differentiated to embryoid bodies. Exp. Cell Res. 229:27-34. - PubMed
-
- Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. - PubMed
-
- Carlson, L. L., A. W. Page, and T. H. Bestor. 1992. Properties and localisation of DNA methyltransferase in pre-implantation mouse embryos: implications for genomic imprinting. Genes Dev. 6:2536-2541. - PubMed
-
- Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
