Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for DNA sequencing and microarrays
- PMID: 15456895
- PMCID: PMC521677
- DOI: 10.1093/nar/gnh127
Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for DNA sequencing and microarrays
Abstract
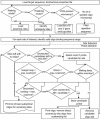
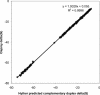
We have developed a software package called Osprey for the calculation of optimal oligonucleotides for DNA sequencing and the creation of microarrays based on either PCR-products or directly spotted oligomers. It incorporates a novel use of position-specific scoring matrices, for the sensitive and specific identification of secondary binding sites anywhere in the target sequence. Using accelerated hardware is faster and more efficient than the traditional pairwise alignments used in most oligo-design software. Osprey consists of a module for target site selection based on user input, novel utilities for dealing with problematic sequences such as repeats, and a common code base for the identification of optimal oligonucleotides from the target list. Overall, these improvements provide a program that, without major increases in run time, reflects current DNA thermodynamics models, improves specificity and reduces the user's data preprocessing and parameterization requirements. Using a TimeLogic hardware accelerator, we report up to 50-fold reduction in search time versus a linear search strategy. Target sites may be derived from computer analysis of DNA sequence assemblies in the case of sequencing efforts, or genome or EST analysis in the case of microarray development in both prokaryotes and eukaryotes.
Figures


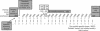

References
-
- Tomiuk S. and Hoffman,K. (2001) Microarray probe selection strategies. Brief. Bioinform., 2, 329–340. - PubMed
-
- Chen B.Y., Janes,H.W. and Chen,S. (2002) Computer programs for PCR primer design and analysis. Methods Mol. Biol., 192, 19–29. - PubMed
-
- Mrowka R., Schuchhardt,J. and Gille,C. (2002) Oligodb—interactive design of oligo DNA for transcription profiling of human genes. Bioinformatics, 18, 1686–1687. - PubMed
-
- Nielsen H.B. and Knudsen,S. (2002) Avoiding cross hybridization by choosing nonredundant targets on cDNA arrays. Bioinformatics, 18, 321–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

