Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa
- PMID: 15458385
- PMCID: PMC1134741
- DOI: 10.1042/BJ20041480
Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa
Abstract
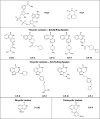
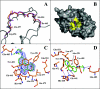
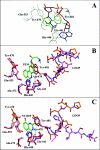
The mono-ADPRT (mono-ADP-ribosyltransferase), Pseudomonas aeruginosa ETA (exotoxin A), catalyses the transfer of ADP-ribose from NAD+ to its protein substrate. A series of water-soluble compounds that structurally mimic the nicotinamide moiety of NAD+ was investigated for their inhibition of the catalytic domain of ETA. The importance of an amide locked into a hetero-ring structure and a core hetero-ring system that is planar was a trend evident by the IC50 values. Also, the weaker inhibitors have core ring structures that are less planar and thus more flexible. One of the most potent inhibitors, PJ34, was further characterized and shown to exhibit competitive inhibition with an inhibition constant K(i) of 140 nM. We also report the crystal structure of the catalytic domain of ETA in complex with PJ34, the first example of a mono-ADPRT in complex with an inhibitor. The 2.1 A (1 A=0.1 nm) resolution structure revealed that PJ34 is bound within the nicotinamide-binding pocket and forms stabilizing hydrogen bonds with the main chain of Gly-441 and to the side-chain oxygen of Gln-485, a member of a proposed catalytic loop. Structural comparison of this inhibitor complex with diphtheria toxin (a mono-ADPRT) and with PARPs [poly(ADP-ribose) polymerases] shows similarity of the catalytic residues; however, a loop similar to that found in ETA is present in diphtheria toxin but not in PARP. The present study provides insight into the important features required for inhibitors that mimic NAD+ and their binding to the mono-ADPRT family of toxins.
Figures

Similar articles
-
Characterization of competitive inhibitors for the transferase activity of Pseudomonas aeruginosa exotoxin A.J Enzyme Inhib Med Chem. 2002 Aug;17(4):235-46. doi: 10.1080/1475636021000010914. J Enzyme Inhib Med Chem. 2002. PMID: 12530476
-
Toward the elucidation of the catalytic mechanism of the mono-ADP-ribosyltransferase activity of Pseudomonas aeruginosa exotoxin A.Biochemistry. 2004 Jan 13;43(1):183-94. doi: 10.1021/bi034772u. Biochemistry. 2004. PMID: 14705944
-
Elucidation of eukaryotic elongation factor-2 contact sites within the catalytic domain of Pseudomonas aeruginosa exotoxin A.Biochem J. 2004 May 1;379(Pt 3):563-72. doi: 10.1042/BJ20031731. Biochem J. 2004. PMID: 14733615 Free PMC article.
-
Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: active-site structure and enzymic mechanism.Curr Top Microbiol Immunol. 1992;175:27-41. doi: 10.1007/978-3-642-76966-5_2. Curr Top Microbiol Immunol. 1992. PMID: 1628498 Review. No abstract available.
-
Stealth and mimicry by deadly bacterial toxins.Trends Biochem Sci. 2006 Feb;31(2):123-33. doi: 10.1016/j.tibs.2005.12.007. Epub 2006 Jan 6. Trends Biochem Sci. 2006. PMID: 16406634 Review.
Cited by
-
PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ß1.PLoS One. 2012;7(6):e37352. doi: 10.1371/journal.pone.0037352. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701565 Free PMC article.
-
Pseudomonas aeruginosa Regulatory Protein AnvM Controls Pathogenicity in Anaerobic Environments and Impacts Host Defense.mBio. 2019 Jul 23;10(4):e01362-19. doi: 10.1128/mBio.01362-19. mBio. 2019. PMID: 31337721 Free PMC article.
-
Molecular insights into plant cell proliferation disturbance by Agrobacterium protein 6b.Genes Dev. 2011 Jan 1;25(1):64-76. doi: 10.1101/gad.1985511. Epub 2010 Dec 14. Genes Dev. 2011. PMID: 21156810 Free PMC article.
-
Knowledge-based fragment binding prediction.PLoS Comput Biol. 2014 Apr 24;10(4):e1003589. doi: 10.1371/journal.pcbi.1003589. eCollection 2014 Apr. PLoS Comput Biol. 2014. PMID: 24762971 Free PMC article.
-
Crystal structures of pertussis toxin with NAD+ and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.J Biol Chem. 2022 May;298(5):101892. doi: 10.1016/j.jbc.2022.101892. Epub 2022 Apr 1. J Biol Chem. 2022. PMID: 35378130 Free PMC article.
References
-
- Lyczak J. B., Cannon C. L., Pier G. B. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. - PubMed
-
- Iglewski B. H., Sadoff J. C. Toxin inhibitors of protein synthesis: production, purification, and assay of Pseudomonas aeruginosa toxin A. Methods Enzymol. 1979;60:780–793. - PubMed
-
- FitzGerald D., Morris R. E., Saelinger C. B. Receptor-mediated internalization of Pseudomonas toxin by mouse fibroblasts. Cell (Cambridge, Mass.) 1980;21:867–873. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

