Determination of thermodynamic parameters for HIV DIS type loop-loop kissing complexes
- PMID: 15459283
- PMCID: PMC521654
- DOI: 10.1093/nar/gkh841
Determination of thermodynamic parameters for HIV DIS type loop-loop kissing complexes
Abstract
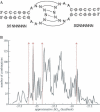
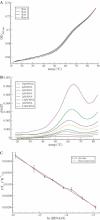
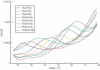
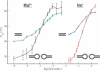
The HIV-1 type dimerization initiation signal (DIS) loop was used as a starting point for the analysis of the stability of Watson-Crick (WC) base pairs in a tertiary structure context. We used ultraviolet melting to determine thermodynamic parameters for loop-loop tertiary interactions and compared them with regular secondary structure RNA helices of the same sequences. In 1 M Na+ the loop-loop interaction of a HIV-1 DIS type pairing is 4 kcal/mol more stable than its sequence in an equivalent regular and isolated RNA helix. This difference is constant and sequence independent, suggesting that the rules governing the stability of WC base pairs in the secondary structure context are also valid for WC base pairs in the tertiary structure context. Moreover, the effect of ion concentration on the stability of loop-loop tertiary interactions differs considerably from that of regular RNA helices. The stabilization by Na+ and Mg2+ is significantly greater if the base pairing occurs within the context of a loop-loop interaction. The dependence of the structural stability on salt concentration was defined via the slope of a T(m)/log [ion] plot. The short base-paired helices are stabilized by 8 degrees C/log [Mg2+] or 11 degrees C/log [Na+], whereas base-paired helices forming tertiary loop-loop interactions are stabilized by 16 degrees C/log [Mg2+] and 26 degrees C/log [Na+]. The different dependence on ionic strength that is observed might reflect the contribution of specific divalent ion binding to the preformation of the hairpin loops poised for the tertiary kissing loop-loop contacts.
Figures
Similar articles
-
Stabilities of HIV-1 DIS type RNA loop-loop interactions in vitro and in vivo.Nucleic Acids Res. 2006 Jan 12;34(1):334-42. doi: 10.1093/nar/gkj435. Print 2006. Nucleic Acids Res. 2006. PMID: 16410613 Free PMC article.
-
Structure and dimerization of HIV-1 kissing loop aptamers.J Mol Biol. 2001 Aug 17;311(3):475-90. doi: 10.1006/jmbi.2001.4879. J Mol Biol. 2001. PMID: 11493002
-
Molecular modeling and dynamics studies of HIV-1 kissing loop structures.J Biomol Struct Dyn. 2002 Dec;20(3):397-412. doi: 10.1080/07391102.2002.10506858. J Biomol Struct Dyn. 2002. PMID: 12437378
-
A structure-based approach for targeting the HIV-1 genomic RNA dimerization initiation site.Biochimie. 2007 Oct;89(10):1195-203. doi: 10.1016/j.biochi.2007.03.003. Epub 2007 Mar 12. Biochimie. 2007. PMID: 17434658 Review.
-
RNA structure at high resolution.FASEB J. 1995 Aug;9(11):1023-33. doi: 10.1096/fasebj.9.11.7544309. FASEB J. 1995. PMID: 7544309 Review.
Cited by
-
Identification of antisense RNA stem-loops that inhibit RNA-protein interactions using a bacterial reporter system.Nucleic Acids Res. 2010 Jun;38(10):3489-501. doi: 10.1093/nar/gkq027. Epub 2010 Feb 15. Nucleic Acids Res. 2010. PMID: 20156995 Free PMC article.
-
RNA helix stability in mixed Na+/Mg2+ solution.Biophys J. 2007 May 15;92(10):3615-32. doi: 10.1529/biophysj.106.100388. Epub 2007 Feb 26. Biophys J. 2007. PMID: 17325014 Free PMC article.
-
Influence of cationic molecules on the hairpin to duplex equilibria of self-complementary DNA and RNA oligonucleotides.Nucleic Acids Res. 2007;35(2):486-94. doi: 10.1093/nar/gkl1073. Epub 2006 Dec 14. Nucleic Acids Res. 2007. PMID: 17169988 Free PMC article.
-
DEAD-box helicase intrinsically disordered domains and structural dynamics of HIV-1 RNA are required to reveal DDX3X catalytic efficiency.Nucleic Acids Res. 2025 Aug 27;53(16):gkaf834. doi: 10.1093/nar/gkaf834. Nucleic Acids Res. 2025. PMID: 40874589 Free PMC article.
-
Interstrand cross-link and bioconjugate formation in RNA from a modified nucleotide.J Org Chem. 2014 Oct 17;79(20):9792-8. doi: 10.1021/jo501982r. Epub 2014 Oct 8. J Org Chem. 2014. PMID: 25295850 Free PMC article.
References
-
- Batey R.T., Rambo,R.P. and Doudna,J.A. (1999) Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl., 38, 2326–2343. - PubMed
-
- Conn G.L. and Draper,D.E. (1998) RNA structure. Curr. Opin. Struct. Biol., 8, 278–285. - PubMed
-
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Szewczak,A.A., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) RNA tertiary structure mediation by adenosine platforms. Science, 273, 1696–1699. - PubMed
-
- Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. - PubMed

