Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta
- PMID: 15459284
- PMCID: PMC521659
- DOI: 10.1093/nar/gkh846
Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta
Abstract
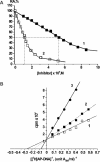
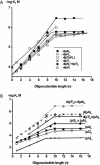
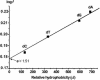
X-ray analysis of enzyme-DNA interactions is very informative in revealing molecular contacts, but provides neither quantitative estimates of the relative importance of these contacts nor information on the relative contributions of specific and nonspecific interactions to the total affinity of enzymes for specific DNA. A stepwise increase in the ligand complexity approach is used to estimate the relative contributions of virtually every nucleotide unit of synthetic DNA containing abasic sites to its affinity for apurinic/apyrimidinic endonuclease (APE1) from human placenta. It was found that APE1 interacts with 9-10 nt units or base pairs of single-stranded and double-stranded ribooligonucleotides and deoxyribooligonucleotides of different lengths and sequences, mainly through weak additive contacts with internucleotide phosphate groups. Such nonspecific interactions of APE1 with nearly every nucleotide within its DNA-binding cleft provides up to seven orders of magnitude (DeltaG degrees approximately -8.7 to -9.0 kcal/mol) of the enzyme affinity for any DNA substrate. In contrast, interactions with the abasic site together with other specific APE1-DNA interactions provide only one order of magnitude (DeltaG degrees approximately -1.1 to -1.5 kcal/mol) of the total affinity of APE1 for specific DNA. We conclude that the enzyme's specificity for abasic sites in DNA is mostly due to a great increase (six to seven orders of magnitude) in the reaction rate with specific DNA, with formation of the Michaelis complex contributing to the substrate preference only marginally.
Figures



Similar articles
-
Thermodynamic, kinetic, and structural basis for recognition and repair of 8-oxoguanine in DNA by Fpg protein from Escherichia coli.Biochemistry. 2002 Jun 18;41(24):7540-8. doi: 10.1021/bi0121297. Biochemistry. 2002. PMID: 12056884
-
Modulation of the Apurinic/Apyrimidinic Endonuclease Activity of Human APE1 and of Its Natural Polymorphic Variants by Base Excision Repair Proteins.Int J Mol Sci. 2020 Sep 28;21(19):7147. doi: 10.3390/ijms21197147. Int J Mol Sci. 2020. PMID: 32998246 Free PMC article.
-
Substrate specificity of human apurinic/apyrimidinic endonuclease APE1 in the nucleotide incision repair pathway.Nucleic Acids Res. 2018 Nov 30;46(21):11454-11465. doi: 10.1093/nar/gky912. Nucleic Acids Res. 2018. PMID: 30329131 Free PMC article.
-
AP Endonuclease 1 as a Key Enzyme in Repair of Apurinic/Apyrimidinic Sites.Biochemistry (Mosc). 2016 Sep;81(9):951-67. doi: 10.1134/S0006297916090042. Biochemistry (Mosc). 2016. PMID: 27682167 Review.
-
Structural, thermodynamic, and kinetic basis for the activities of some nucleic acid repair enzymes.J Mol Recognit. 2011 Jul-Aug;24(4):656-77. doi: 10.1002/jmr.1096. J Mol Recognit. 2011. PMID: 21584877 Review.
Cited by
-
APE1: A skilled nucleic acid surgeon.DNA Repair (Amst). 2018 Nov;71:93-100. doi: 10.1016/j.dnarep.2018.08.012. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30170830 Free PMC article. Review.
-
[Homologous DNA transferase RecA: functional activities and the search for homology by recombining DNA molecules].Mol Biol (Mosk). 2007 May-Jun;41(3):467-77. doi: 10.1134/s0026893307030132. Mol Biol (Mosk). 2007. PMID: 17685224 Review. Russian.
-
Role of active site tyrosines in dynamic aspects of DNA binding by AP endonuclease.DNA Repair (Amst). 2007 Mar 1;6(3):374-82. doi: 10.1016/j.dnarep.2006.11.004. Epub 2007 Jan 10. DNA Repair (Amst). 2007. PMID: 17218168 Free PMC article.
-
APE1 is dispensable for S-region cleavage but required for its repair in class switch recombination.Proc Natl Acad Sci U S A. 2014 Dec 2;111(48):17242-7. doi: 10.1073/pnas.1420221111. Epub 2014 Nov 17. Proc Natl Acad Sci U S A. 2014. PMID: 25404348 Free PMC article.
-
Diffusion within the cytoplasm: a mesoscale model of interacting macromolecules.Biophys J. 2014 Dec 2;107(11):2579-91. doi: 10.1016/j.bpj.2014.09.043. Epub 2014 Dec 2. Biophys J. 2014. PMID: 25468337 Free PMC article.
References
-
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
-
- Mol C.D., Hosfield,D.J. and Tainer,J.A. (2000) Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res., 460, 211–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

