A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys
- PMID: 15459285
- PMCID: PMC521638
- DOI: 10.1093/nar/gkh785
A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys
Abstract
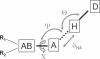
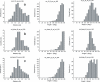
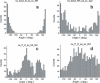
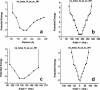
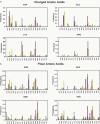
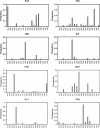
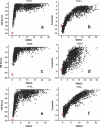
RNA-binding proteins play many essential roles in the regulation of gene expression in the cell. Despite the significant increase in the number of structures for RNA-protein complexes in the last few years, the molecular basis of specificity remains unclear even for the best-studied protein families. We have developed a distance and orientation-dependent hydrogen-bonding potential based on the statistical analysis of hydrogen-bonding geometries that are observed in high-resolution crystal structures of protein-DNA and protein-RNA complexes. We observe very strong geometrical preferences that reflect significant energetic constraints on the relative placement of hydrogen-bonding atom pairs at protein-nucleic acid interfaces. A scoring function based on the hydrogen-bonding potential discriminates native protein-RNA structures from incorrectly docked decoys with remarkable predictive power. By incorporating the new hydrogen-bonding potential into a physical model of protein-RNA interfaces with full atom representation, we were able to recover native amino acids at protein-RNA interfaces.
Figures


Similar articles
-
A knowledge-based potential function predicts the specificity and relative binding energy of RNA-binding proteins.FEBS J. 2007 Dec;274(24):6378-91. doi: 10.1111/j.1742-4658.2007.06155.x. Epub 2007 Nov 12. FEBS J. 2007. PMID: 18005254
-
Knowledge-based scoring functions in drug design: 3. A two-dimensional knowledge-based hydrogen-bonding potential for the prediction of protein-ligand interactions.J Chem Inf Model. 2011 Nov 28;51(11):2994-3004. doi: 10.1021/ci2003939. Epub 2011 Oct 28. J Chem Inf Model. 2011. PMID: 21999432
-
Structure-based analysis of protein-RNA interactions using the program ENTANGLE.J Mol Biol. 2001 Aug 3;311(1):75-86. doi: 10.1006/jmbi.2001.4857. J Mol Biol. 2001. PMID: 11469858
-
Themes in RNA-protein recognition.J Mol Biol. 1999 Oct 22;293(2):255-70. doi: 10.1006/jmbi.1999.2991. J Mol Biol. 1999. PMID: 10550207 Review.
-
Targeting of nucleic acids by iron complexes.Met Ions Biol Syst. 1996;33:453-84. Met Ions Biol Syst. 1996. PMID: 8742852 Review. No abstract available.
Cited by
-
Kernel-based machine learning protocol for predicting DNA-binding proteins.Nucleic Acids Res. 2005 Nov 10;33(20):6486-93. doi: 10.1093/nar/gki949. Print 2005. Nucleic Acids Res. 2005. PMID: 16284202 Free PMC article.
-
Bioinformatics Tools and Benchmarks for Computational Docking and 3D Structure Prediction of RNA-Protein Complexes.Genes (Basel). 2018 Aug 25;9(9):432. doi: 10.3390/genes9090432. Genes (Basel). 2018. PMID: 30149645 Free PMC article. Review.
-
Engineering RNA-binding proteins for biology.FEBS J. 2013 Aug;280(16):3734-54. doi: 10.1111/febs.12375. Epub 2013 Jul 5. FEBS J. 2013. PMID: 23742071 Free PMC article. Review.
-
Structure-based prediction of C2H2 zinc-finger binding specificity: sensitivity to docking geometry.Nucleic Acids Res. 2007;35(4):1085-97. doi: 10.1093/nar/gkl1155. Epub 2007 Jan 30. Nucleic Acids Res. 2007. PMID: 17264128 Free PMC article.
-
Assessing the performance of MM/PBSA and MM/GBSA methods. 8. Predicting binding free energies and poses of protein-RNA complexes.RNA. 2018 Sep;24(9):1183-1194. doi: 10.1261/rna.065896.118. Epub 2018 Jun 21. RNA. 2018. PMID: 29930024 Free PMC article.
References
-
- Wolfe S.A., Nekludova,L. and Pabo,C.O. (2000) DNA recognition by Cys(2)His(2) zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct., 29, 183–212. - PubMed
-
- Jamieson A.C., Miller,J.C. and Pabo,C.O. (2003) Drug discovery with engineered zinc-finger proteins. Nature Rev. Drug Discov., 2, 361–368. - PubMed
-
- Laird-Offringa I.A. and Belasco,J.G. (1998) RNA-binding proteins tamed. Nature Struct. Biol., 5, 665–668. - PubMed

