Effect of sodium dodecyl sulfate on folding and thermal stability of acid-denatured cytochrome c: a spectroscopic approach
- PMID: 15459332
- PMCID: PMC2286590
- DOI: 10.1110/ps.04827604
Effect of sodium dodecyl sulfate on folding and thermal stability of acid-denatured cytochrome c: a spectroscopic approach
Abstract
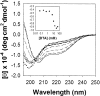
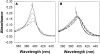
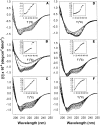
The molten globule (MG) state can be an intermediate in the protein folding pathway; thus, its detailed description can help understanding protein folding. Sodium dodecyl sulfate (SDS), an anionic surfactant that is commonly used to mimic hydrophobic binding environments such as cell membranes, is known to denature some native state proteins, including horse cytochrome c (cyt c). In this article, refolding of acid denatured cyt c is studied under the influence of SDS to form MG-like states at both low concentration and above the critical micelle concentration using Fourier transform Infrared (FTIR) and ultraviolet and visible absorption as well as fluorescence and circular dichroism (CD). Thermal denaturation monitored with FTIR and CD shows distinct final high temperature states starting from MG-like states formed with different SDS/protein ratios. The results suggest that the SDS/protein ratio as well as the actual SDS (or protein) concentration affects structure and its thermal stability. Thermal denaturation monitored with CD and FTIR for cyt c at neutral pH but denatured with SDS showed that at a high SDS/protein ratio, the thermal behavior of MG-like states formed at low and neutral pH are quite similar. Based on the results obtained, the merits of two models of the protein-surfactant structure are discussed for different SDS concentrations.
Figures




References
-
- Brems, D.N., and Stellwagen, E. 1983. Manipulate of the observed kinetics phases in the refolding of denatured ferricytochromes c. J. Biol. Chem. 258 3655–3660. - PubMed
-
- Bushnell, G.W., Louie, G.V., and Brayer, G.D. 1990. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 214 585–595. - PubMed
-
- Chattopadhyay, K. and Mazumdar, S. 2003. Stabilization of partially folded states of cytochrome c in aqueous surfactant: Effects of ionic and hydrophobic interactions. Biochemistry 42 14606–14613. - PubMed
-
- Chen, S.H. and Teixeira, J. 1986. Structure and fractal dimension of protein–detergent complexes. Phys. Rev. Lett. 57 2583–2586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

