Molecular mechanisms of translational control
- PMID: 15459663
- PMCID: PMC7097087
- DOI: 10.1038/nrm1488
Molecular mechanisms of translational control
Abstract
Translational control is widely used to regulate gene expression. This mode of regulation is especially relevant in situations where transcription is silent or when local control over protein accumulation is required. Although many examples of translational regulation have been described, only a few are beginning to be mechanistically understood. Instead of providing a comprehensive account of the examples that are known at present, we discuss instructive cases that serve as paradigms for different modes of translational control.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
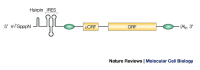
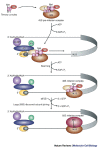
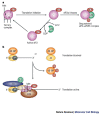
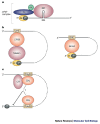
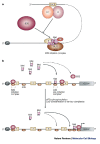
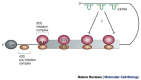
References
-
- Wickens M, Goodwin E, Kimble. J, Strickland S, Hentze M. Translational control of gene expression. 2000. p. 295.
-
- Hershey JWB, Merrick WC. Translational control of gene expression. 2000. p. 33.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

