Metabolic control analysis under uncertainty: framework development and case studies
- PMID: 15465856
- PMCID: PMC1304888
- DOI: 10.1529/biophysj.104.048090
Metabolic control analysis under uncertainty: framework development and case studies
Abstract
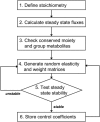
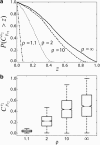
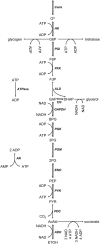
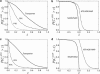
Information about the enzyme kinetics in a metabolic network will enable understanding of the function of the network and quantitative prediction of the network responses to genetic and environmental perturbations. Despite recent advances in experimental techniques, such information is limited and existing experimental data show extensive variation and they are based on in vitro experiments. In this article, we present a computational framework based on the well-established (log)linear formalism of metabolic control analysis. The framework employs a Monte Carlo sampling procedure to simulate the uncertainty in the kinetic data and applies statistical tools for the identification of the rate-limiting steps in metabolic networks. We applied the proposed framework to a branched biosynthetic pathway and the yeast glycolysis pathway. Analysis of the results allowed us to interpret and predict the responses of metabolic networks to genetic and environmental changes, and to gain insights on how uncertainty in the kinetic mechanisms and kinetic parameters propagate into the uncertainty in predicting network responses. Some of the practical applications of the proposed approach include the identification of drug targets for metabolic diseases and the guidance for design strategies in metabolic engineering for the purposeful manipulation of the metabolism of industrial organisms.
Figures




References
-
- Almaas, E., B. Kovacs, T. Vicsek, Z. N. Oltvai, and A. L. Barabasi. 2004. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 427:839–843. - PubMed
-
- Alves, R., and M. A. Savageau. 2000. Systemic properties of ensembles of metabolic networks: application of graphical and statistical methods to simple unbranched pathways. Bioinformatics. 16:534–547. - PubMed
-
- Andersen, K. B., and K. von Meyenburg. 1977. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J. Biol. Chem. 252:4151–4156. - PubMed
-
- Atkinson, D. E. 1968. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 7:4030–4034. - PubMed
-
- Bailey, J. E. 1991. Toward a science of metabolic engineering. Science. 252:1668–1675. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

