Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development
- PMID: 15465908
- PMCID: PMC522052
- DOI: 10.1073/pnas.0406451101
Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development
Abstract
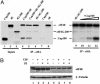
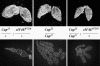
In Drosophila, the product of the fs (2)cup gene (Cup) is known to be crucial for diverse aspects of female germ-line development. Its functions at the molecular level, however, have remained mainly unexplored. Cup was found to directly associate with eukaryotic translation initiation factor 4E (eIF4E). In this report, we show that Cup is a nucleocytoplasmic shuttling protein and that the interaction with eIF4E promotes retention of the Cup protein in the cytoplasm. Cup is required for the correct accumulation and localization of eIF4E within the posterior cytoplasm of developing oocytes. We furthermore show that cup and eIF4E interact genetically, because a reduction in the level of eIF4E activity deteriorates the development and growth of ovaries bearing homozygous cup mutant alleles. Our results reveal a crucial role for the Cup-eIF4E complex in ovary-specific developmental programs.
Figures
Similar articles
-
Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis.Dev Cell. 2004 Jan;6(1):69-78. doi: 10.1016/s1534-5807(03)00400-3. Dev Cell. 2004. PMID: 14723848
-
Mextli is a novel eukaryotic translation initiation factor 4E-binding protein that promotes translation in Drosophila melanogaster.Mol Cell Biol. 2013 Aug;33(15):2854-64. doi: 10.1128/MCB.01354-12. Epub 2013 May 28. Mol Cell Biol. 2013. PMID: 23716590 Free PMC article.
-
Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz.J Cell Biol. 2003 Dec 22;163(6):1197-204. doi: 10.1083/jcb.200309088. J Cell Biol. 2003. PMID: 14691132 Free PMC article.
-
A cup full of functions.RNA Biol. 2005 Oct-Dec;2(4):125-8. doi: 10.4161/rna.2.4.2416. Epub 2005 Dec 14. RNA Biol. 2005. PMID: 17114932 Review.
-
Contrasting mechanisms of regulating translation of specific Drosophila germline mRNAs at the level of 5'-cap structure binding.Biochem Soc Trans. 2005 Dec;33(Pt 6):1544-6. doi: 10.1042/BST0331544. Biochem Soc Trans. 2005. PMID: 16246166 Review.
Cited by
-
4E-T-bound mRNAs are stored in a silenced and deadenylated form.Genes Dev. 2020 Jun 1;34(11-12):847-860. doi: 10.1101/gad.336073.119. Epub 2020 Apr 30. Genes Dev. 2020. PMID: 32354837 Free PMC article.
-
Translational Control during Developmental Transitions.Cold Spring Harb Perspect Biol. 2019 Jun 3;11(6):a032987. doi: 10.1101/cshperspect.a032987. Cold Spring Harb Perspect Biol. 2019. PMID: 30082467 Free PMC article. Review.
-
eIF4E: new family members, new binding partners, new roles.J Biol Chem. 2009 Jun 19;284(25):16711-16715. doi: 10.1074/jbc.R900002200. Epub 2009 Feb 23. J Biol Chem. 2009. PMID: 19237539 Free PMC article. Review.
-
Cup blocks the precocious activation of the orb autoregulatory loop.PLoS One. 2011;6(12):e28261. doi: 10.1371/journal.pone.0028261. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164257 Free PMC article.
-
Regulation of Germ Cell mRNPs by eIF4E:4EIP Complexes: Multiple Mechanisms, One Goal.Front Cell Dev Biol. 2020 Jul 7;8:562. doi: 10.3389/fcell.2020.00562. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32733883 Free PMC article. Review.
References
-
- Spradling, A. C. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Arias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. I, pp. 1–70.
-
- Keyes, L. N. & Spradling, A. C. (1997) Development (Cambridge, U.K.) 124, 1419–1431. - PubMed
-
- Verrotti, A. C. & Wharton, R. P. (2000) Development (Cambridge, U.K.) 127, 5225–5232. - PubMed
-
- Dahanukar, A., Walker, J. A. & Wharton, R. P. (1999) Mol. Cell 4, 209–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

