MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli
- PMID: 15466019
- PMCID: PMC522180
- DOI: 10.1128/JB.186.20.6689-6697.2004
MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli
Abstract
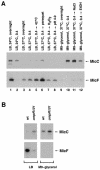
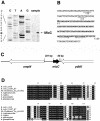
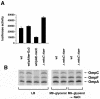
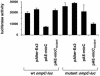
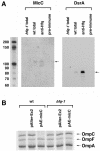
In a previous bioinformatics-based search for novel small-RNA genes encoded by the Escherichia coli genome, we identified a region, IS063, located between the ompN and ydbK genes, that encodes an approximately 100-nucleotide small-RNA transcript. Here we show that the expression of this small RNA is increased at a low temperature and in minimal medium. Twenty-two nucleotides at the 5' end of this transcript have the potential to form base pairs with the leader sequence of the mRNA encoding the outer membrane protein OmpC. The deletion of IS063 increased the expression of an ompC-luc translational fusion 1.5- to 2-fold, and a 10-fold overexpression of the small RNA led to a 2- to 3-fold repression of the fusion. Deletion and overexpression of the IS063 RNA also resulted in increases and decreases, respectively, in OmpC protein levels. Taken together, these results suggest that IS063 is a regulator of OmpC expression; thus, the small RNA has been renamed MicC. The antisense regulation was further demonstrated by the finding that micC mutations were suppressed by compensatory mutations in the ompC mRNA. MicC was also shown to inhibit ribosome binding to the ompC mRNA leader in vitro and to require the Hfq RNA chaperone for its function. We suggest that the MicF and MicC RNAs act in conjunction with the EnvZ-OmpR two-component system to control the OmpF/OmpC protein ratio in response to a variety of environmental stimuli.
Figures

Comment in
-
Regulatory small RNAs: the key to coordinating global regulatory circuits.J Bacteriol. 2004 Oct;186(20):6679-80. doi: 10.1128/JB.186.20.6679-6680.2004. J Bacteriol. 2004. PMID: 15466017 Free PMC article. No abstract available.
References
-
- Altuvia, S., D. Weinsterin-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. - PubMed
-
- Andersen, J., S. A. Forst, K. Zhao, M. Inouye, and N. Delihas. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961-17970. - PubMed
-
- Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

