GadY, a small-RNA regulator of acid response genes in Escherichia coli
- PMID: 15466020
- PMCID: PMC522195
- DOI: 10.1128/JB.186.20.6698-6705.2004
GadY, a small-RNA regulator of acid response genes in Escherichia coli
Abstract
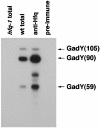
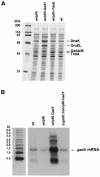
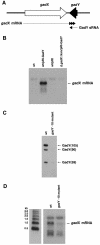
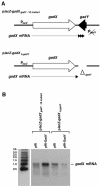
A previous bioinformatics-based search for small RNAs in Escherichia coli identified a novel RNA named IS183. The gene encoding this small RNA is located between and on the opposite strand of genes encoding two transcriptional regulators of the acid response, gadX (yhiX) and gadW (yhiW). Given that IS183 is encoded in the gad gene cluster and because of its role in regulating acid response genes reported here, this RNA has been renamed GadY. We show that GadY exists in three forms, a long form consisting of 105 nucleotides and two processed forms, consisting of 90 and 59 nucleotides. The expression of this small RNA is highly induced during stationary phase in a manner that is dependent on the alternative sigma factor sigmaS. Overexpression of the three GadY RNA forms resulted in increased levels of the mRNA encoding the GadX transcriptional activator, which in turn caused increased levels of the GadA and GadB glutamate decarboxylases. A promoter mutation which abolished gadY expression resulted in a reduction in the amount of gadX mRNA during stationary phase. The gadY gene was shown to overlap the 3' end of the gadX gene, and this overlap region was found to be necessary for the GadY-dependent accumulation of gadX mRNA. We suggest that during stationary phase, GadY forms base pairs with the 3'-untranslated region of the gadX mRNA and confers increased stability, allowing for gadX mRNA accumulation and the increased expression of downstream acid resistance genes.
Figures


Comment in
-
Regulatory small RNAs: the key to coordinating global regulatory circuits.J Bacteriol. 2004 Oct;186(20):6679-80. doi: 10.1128/JB.186.20.6679-6680.2004. J Bacteriol. 2004. PMID: 15466017 Free PMC article. No abstract available.
References
-
- Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300:1101-1112. - PubMed
-
- Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. - PubMed
-
- Bordi, C., L. Theraulaz, V. Mejean, and C. Jourlin-Castelli. 2003. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 48:211-223. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

