ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth
- PMID: 15466051
- PMCID: PMC522188
- DOI: 10.1128/JB.186.20.6983-6998.2004
ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth
Abstract
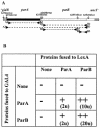
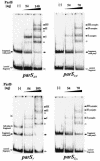
The par genes of Pseudomonas aeruginosa have been studied to increase the understanding of their mechanism of action and role in the bacterial cell. Key properties of the ParB protein have been identified and are associated with different parts of the protein. The ParB- ParB interaction domain was mapped in vivo and in vitro to the C-terminal 56 amino acids (aa); 7 aa at the C terminus play an important role. The dimerization domain of P. aeruginosa ParB is interchangeable with the dimerization domain of KorB from plasmid RK2 (IncP1 group). The C-terminal part of ParB is also involved in ParB-ParA interactions. Purified ParB binds specifically to DNA containing a putative parS sequence based on the consensus sequence found in the chromosomes of Bacillus subtilis, Pseudomonas putida, and Streptomyces coelicolor. The overproduction of ParB was shown to inhibit the function of genes placed near parS. This "silencing" was dependent on the parS sequence and its orientation. The overproduction of P. aeruginosa ParB or its N-terminal part also causes inhibition of the growth of P. aeruginosa and P. putida but not Escherichia coli cells. Since this inhibitory determinant is located well away from ParB segments required for dimerization or interaction with the ParA counterpart, this result may suggest a role for the N terminus of P. aeruginosa ParB in interactions with host cell components.
Figures





References
-
- Arnold, F. H. 1991. Metal-affinity separations: a new dimension in protein processing. Bio/Technology 9:151-156. - PubMed
-
- Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. - PubMed
-
- Bignell, C. R., A. S. Haines, D. Khare, and C. M. Thomas. 1999. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 34:205-216. - PubMed
-
- Bignell, C. R., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

