Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis
- PMID: 15466233
- PMCID: PMC523374
- DOI: 10.1104/pp.104.046169
Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis
Abstract
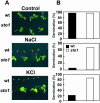
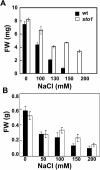
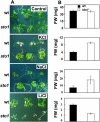
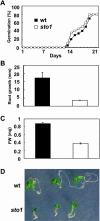
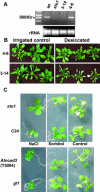
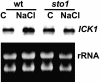
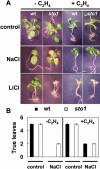
We have identified a T-DNA insertion mutation of Arabidopsis (ecotype C24), named sto1 (salt tolerant), that results in enhanced germination on both ionic (NaCl) and nonionic (sorbitol) hyperosmotic media. sto1 plants were more tolerant in vitro than wild type to Na(+) and K(+) both for germination and subsequent growth but were hypersensitive to Li(+). Postgermination growth of the sto1 plants on sorbitol was not improved. Analysis of the amino acid sequence revealed that STO1 encodes a 9-cis-epoxicarotenoid dioxygenase (similar to 9-cis-epoxicarotenoid dioxygenase GB:AAF26356 [Phaseolus vulgaris] and to NCED3 GB:AB020817 [Arabidopsis]), a key enzyme in the abscisic acid (ABA) biosynthetic pathway. STO1 transcript abundance was substantially reduced in mutant plants. Mutant sto1 plants were unable to accumulate ABA following a hyperosmotic stress, although their basal ABA level was only moderately altered. Either complementation of the sto1 with the native gene from the wild-type genome or supplementation of ABA to the growth medium restored the wild-type phenotype. Improved growth of sto1 mutant plants on NaCl, but not sorbitol, medium was associated with a reduction in both NaCl-induced expression of the ICK1 gene and ethylene accumulation. Osmotic adjustment of sto1 plants was substantially reduced compared to wild-type plants under conditions where sto1 plants grew faster. The sto1 mutation has revealed that reduced ABA can lead to more rapid growth during hyperionic stress by a signal pathway that apparently is at least partially independent of signals that mediate nonionic osmotic responses.
Figures






References
-
- Bray EA (2002. b) Abscisic acid regulation of gene expression during-water deficit stress in the era of the Arabidopsis genome. Plant Cell Environ 25: 153–161 - PubMed
-
- Bressan RA, Nelson DE, Iraki NM, LaRosa PC, Singh NK, Hasegawa PM, Carpita NC (1990) Reduced cell expansion and changes in cell walls of plant cells adapted to NaCl. In F Katterman, ed, Environmental Injury to Plants. Academic Press, San Diego, pp 137–171
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

