Comparative sequence analysis of the region harboring the hardness locus in barley and its colinear region in rice
- PMID: 15466237
- PMCID: PMC523377
- DOI: 10.1104/pp.104.044081
Comparative sequence analysis of the region harboring the hardness locus in barley and its colinear region in rice
Abstract
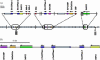
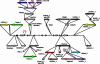
The ancestral shared synteny concept has been advocated as an approach to positionally clone genes from complex genomes. However, the unified grass genome model and the study of grasses as a single syntenic genome is a topic of considerable controversy. Hence, more quantitative studies of cereal colinearity at the sequence level are required. This study compared a contiguous 300-kb sequence of the barley (Hordeum vulgare) genome with the colinear region in rice (Oryza sativa). The barley sequence harbors genes involved in endosperm texture, which may be the subject of distinctive evolutionary forces and is located at the extreme telomeric end of the short arm of chromosome 5H. Comparative sequence analysis revealed the presence of five orthologous genes and a complex, postspeciation evolutionary history involving small chromosomal rearrangements, a translocation, numerous gene duplications, and extensive transposon insertion. Discrepancies in gene content and microcolinearity indicate that caution should be exercised in the use of rice as a surrogate for map-based cloning of genes from large genome cereals such as barley.
Figures


References
-
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 211–215
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

