Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes
- PMID: 15466286
- PMCID: PMC524409
- DOI: 10.1101/gr.2542904
Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes
Abstract
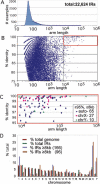
We have performed the first genome-wide analysis of the Inverted Repeat (IR) structure in the human genome, using a novel and efficient software package called Inverted Repeats Finder (IRF). After masking of known repetitive elements, IRF detected 22,624 human IRs characterized by arm size from 25 bp to >100 kb with at least 75% identity, and spacer length up to 100 kb. This analysis required 6 h on a desktop PC. In all, 166 IRs had arm lengths >8 kb. From this set, IRs were excluded if they were in unfinished/unassembled regions of the genome, or clustered with other closely related IRs, yielding a set of 96 large IRs. Of these, 24 (25%) occurred on the X-chromosome, although it represents only approximately 5% of the genome. Of the X-chromosome IRs, 83.3% were >/=99% identical, compared with 28.8% of autosomal IRs. Eleven IRs from Chromosome X, one from Chromosome 11, and seven already described from Chromosome Y contain genes predominantly expressed in testis. PCR analysis of eight of these IRs correctly amplified the corresponding region in the human genome, and six were also confirmed in gorilla or chimpanzee genomes. Similarity dot-plots revealed that 22 IRs contained further secondary homologous structures partially categorized into three distinct patterns. The prevalence of large highly homologous IRs containing testes genes on the X- and Y-chromosomes suggests a possible role in male germ-line gene expression and/or maintaining sequence integrity by gene conversion.
Figures



References
-
- Aradhya, S., Bardaro, T., Galgoczy, P., Yamagata, T., Esposito, T., Patlan, H., Ciccodicola, A., Munnich, A., Kenwrick, S., Platzer, M., et al. 2001. Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum. Mol. Genet. 10: 2557-2567. - PubMed
-
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. - PubMed
-
- Benham, C.J., Savitt, A.G., and Bauer, W.R. 2002. Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: Complete determination of energetics using a statistical mechanical model. J. Mol. Biol. 316: 563-581. - PubMed
WEB SITE REFERENCES
-
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
-
- http://tandem.bu.edu/cgi-bin/irdb/irdb.exe; Inverted Repeat Finder.
-
- http://tandem.bu.edu; Inverted Repeat Data Base (IRDB).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
