Satellite DNA from the Y chromosome of the malaria vector Anopheles gambiae
- PMID: 15466420
- PMCID: PMC1448884
- DOI: 10.1534/genetics.104.034264
Satellite DNA from the Y chromosome of the malaria vector Anopheles gambiae
Abstract
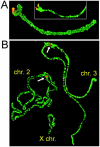
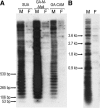
Satellite DNA is an enigmatic component of genomic DNA with unclear function that has been regarded as "junk." Yet, persistence of these tandem highly repetitive sequences in heterochromatic regions of most eukaryotic chromosomes attests to their importance in the genome. We explored the Anopheles gambiae genome for the presence of satellite repeats and identified 12 novel satellite DNA families. Certain families were found in close juxtaposition within the genome. Six satellites, falling into two evolutionarily linked groups, were investigated in detail. Four of them were experimentally confirmed to be linked to the Y chromosome, whereas their relatives occupy centromeric regions of either the X chromosome or the autosomes. A complex evolutionary pattern was revealed among the AgY477-like satellites, suggesting their rapid turnover in the A. gambiae complex and, potentially, recombination between sex chromosomes. The substitution pattern suggested rolling circle replication as an array expansion mechanism in the Y-linked 53-bp satellite families. Despite residing in different portions of the genome, the 53-bp satellites share the same monomer lengths, apparently maintained by molecular drive or structural constraints. Potential functional centromeric DNA structures, consisting of twofold dyad symmetries flanked by a common sequence motif, have been identified in both satellite groups.
Figures




References
-
- Besansky, N. J., and J. R. Powell, 1992. Reassociation kinetics of Anopheles gambiae (Diptera: Culicidae) DNA. J. Med. Entomol. 29: 125–128. - PubMed
-
- Biessmann, H., J. Donath and M. F. Walter, 1996. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol. Biol. 5: 11–20. - PubMed
-
- Bonaccorsi, S., G. Santini, M. Gatti, S. Pimpinelli and M. Colluzzi, 1980. Intraspecific polymorphism of sex chromosome heterochromatin in two species of the Anopheles gambiae complex. Chromosoma 76: 57–64. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

